Genetic Testing for Cystic Fibrosis - CAM 044
Description
Cystic fibrosis (CF) is an autosomal recessive genetic disease in which dysfunctional epithelial chloride channels lead to excessively thick mucus affecting multiple organ systems. Common complications include mucous plugging of the airway, lung inflammation, chronic pulmonary infections, intestinal malabsorption, pancreatic insufficiency, and infertility.1
When pursuing genetic testing for cystic fibrosis, genetic counseling is strongly recommended.
For guidance on pre-implantation genetic testing, please see CAM 110-Pre-Implantation Genetic Testing. For guidance on prenatal screening and preconception screening for cystic fibrosis, please see CAM-358-Prenatal Screening (Genetic).
Policy
Application of coverage criteria is dependent upon an individual’s benefit coverage at the time of the request.
- Common variant testing (see Note 1) or comprehensive analysis of the CFTR gene (i.e., full gene sequencing, deletion/duplication analysis) is considered MEDICALLY NECESSARY in any of the following situations:
- In a newborn after an abnormal newborn screening result using immunoreactive trypsinogen.
- For infants with meconium ileus.
- For individuals with congenital absence of the vas deferens (CAVD).
- For individuals with idiopathic pancreatitis or bronchiectasis.
- As an adjunct to sweat testing in an individual presenting with symptoms of cystic fibrosis, common variant testing (see Note 1) or comprehensive analysis of the CFTR gene (i.e., full gene sequencing, deletion/duplication analysis) is considered MEDICALLY NECESSARY in any of the following situations:
- When there are no known familial pathogenic or likely pathogenic (P/LP) variants.
- When one or more familial P/LP variants are known; testing must include the familial P/LP variants.
- For individuals with congenital bilateral absence of the vas deferens (CBAVD), common variant testing (see Note 1) or comprehensive analysis of the CFTR gene (i.e., full gene sequencing, deletion/duplication analysis), including testing for the CFTR IVS8 5T/7T/9T variant, is considered MEDICALLY NECESSARY.
- When the CFTR R117H variant is detected on carrier screening, reflex genetic testing for the CFTR IVS8 5T/7T/9T variant is considered MEDICALLY NECESSARY.
- For individuals for whom common variant testing (see Note 1) identified no variants or only one variant, but for whom the clinical suspicion of cystic fibrosis still remains, comprehensive analysis of the CFTR gene (i.e., full gene sequencing, deletion/duplication analysis) is considered MEDICALLY NECESSARY in any of the following situations:
- For individuals that presented with symptoms of cystic fibrosis and who received panel testing as an adjunct to sweat testing.
- When a diagnosis of cystic fibrosis related CBAVD remains a consideration in individuals with CBAVD.
- For the family members of individuals with CFTR-related metabolic disorder/cystic fibrosis screen positive, inconclusive diagnosis (CRMS/CFSPID), comprehensive analysis of the CFTR gene (i.e., full gene sequencing, deletion/duplication analysis) is considered MEDICALLY NECESSARY in any of the following situations:
- For parents of the individual: When phasing of the CFTR variants would inform the diagnostic status of the individuals by confirming the inheritance pattern.
- For siblings of the individual.
- For all other situations not described above, genetic testing for variants in the CFTR gene is considered NOT MEDICALLY NECESSARY.
NOTES:
Note 1: Common variant testing for CFTR mutations must include the American College of Medical Genetics’ (ACMG) CFTR carrier screening variant set (n = 100). Please see the “Guidelines and Recommendations” section of this policy for a table of ACMG’s CFTR minimum variant set.
Table of Terminology
| Term |
Definition |
| ACMG |
American College of Medical Genetics |
| ACOG |
American College of Obstetrics and Gynecology |
| CAVD |
Congenital absence of the vas deferens |
| CBAVD |
Congenital bilateral absence of the vas deferens |
| CF |
Cystic fibrosis |
| CFCD |
Cystic fibrosis carrier detection |
| CFF |
Cystic Fibrosis Foundation |
| CFSPID |
Cystic fibrosis screen positive, inconclusive diagnosis |
| CFTR |
Cystic fibrosis transmembrane conductance regulator |
| CFTR2 |
Clinical and functional translation of CFTR |
| CLIA ’88 |
Clinical Laboratory Improvement Amendments Of 1988 |
| CLSI |
Clinical and Laboratory Standards Institute |
| CMS |
Centers For Medicare and Medicaid |
| CPIC |
Clinical Pharmacogenetics Implementation Consortium |
| CRMS |
CFTR-related metabolic disorder |
| CVS |
Chorionic villus sampling |
| DNA |
Deoxyribonucleic acid |
| ECFS |
European Cystic Fibrosis Society |
| FDA |
Food and Drug Administration |
| FEV1 |
Forced expiratory volume in 1 second |
| ICM |
Intestinal current measurement |
| IRT |
Immunoreactive trypsinogen |
| IVA |
Ivacaftor |
| LDT |
Laboratory-developed test |
| LUM |
Lumacaftor |
| MMWR |
Morbidity And Mortality Weekly Report |
| MVCC |
Mutation of varying clinical consequence |
| NBS |
Newborn screening |
| NGS |
Next generation sequencing |
| NICE |
National Institute for Health and Care Excellence |
| NPD |
Nasal potential difference |
| NSGC |
National Society of Genetic Counselors |
| P/LP |
Pathogenic/likely pathogenic |
| PCR |
Polymerase chain reaction |
| PCRM |
Physicians Committee for Responsible Medicine |
| PCS |
Cystic fibrosis carrier screening |
| QALY |
Quality adjusted life year |
| RT-PCR |
Reverse transcription polymerase chain reaction |
| TDAS |
XTAG Data Analysis Software |
| TDN |
Therapeutics Development Network |
| TOP |
Termination of pregnancy |
| UNK |
Uncharacterized mutation |
| VUS |
Variant of uncertain significance |
Rationale
Cystic fibrosis (CF) is a common life-limiting autosomal recessive genetic disorder caused by the mutation of a gene that encodes an epithelial chloride-conducting transmembrane channel called the cystic fibrosis transmembrane conductance regulator (CFTR). This gene regulates anion (negatively charged ion) transport and mucociliary clearance. Mucociliary clearance is the process by which particles and gases dissolved in the mucus are moved unidirectionally from the respiratory tract. The CFTR protein was first identified as a chloride channel but has been shown to facilitate or regulate the transport of other ions, such as sodium, thiocyanate, bicarbonate, as well as water absorption and excretion.2,3 The CFTR protein is present in the epithelia of various tissues, including that of the lungs, sweat glands, gastrointestinal tract, and pancreas.4 CFTR dysfunction mainly affects epithelial cells; although, there is evidence of a role in immune cells.
Mutations in the CFTR gene which impede protein production, stability, or activity result in less available functional protein.3 Functional failure of CFTR results in defective mucociliary clearance, chronic infection, and abnormal inflammatory response leading to progressive, irreversible lung damage.2 Other manifestations of dysfunctional CFTR include pancreatic insufficiency and meconium ileus.4 The early identification and treatment of patients by multidisciplinary teams have resulted in improvements in both quality of life and clinical outcomes in patients with cystic fibrosis, with life expectancy reaching 50 or even 60 years.3
Most mutations of the CFTR gene are missense alterations, but other mutations, deletions and insertions have been described.5 Missense mutations occur when a single nucleotide is changed, resulting in a different amino acid which may affect the overall protein functionality. To date, over 2000 mutations have been identified in the CFTR gene.6 CFTR mutations can be divided into six classes according to their effects on protein function as depicted in the figure below.5
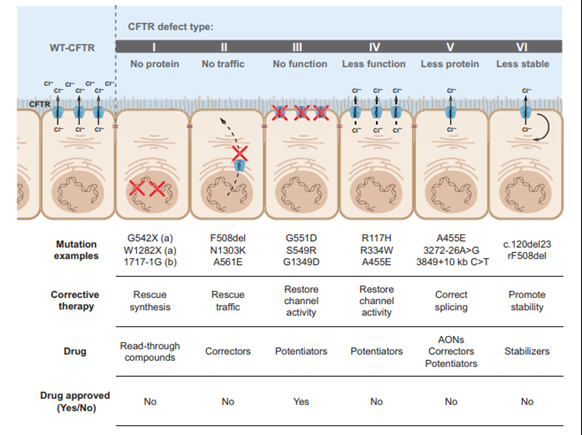
Class I, II, and III mutations are associated with no residual CFTR function and patients with these mutations have a severe phenotype, whereas individuals with class IV, V, and VI mutations have some residual function of CFTR protein and have a mild lung phenotype and pancreatic sufficiency.5,7
Due to the relatively high frequency in the U.S. population and studies demonstrating that early detection and care can improve outcomes, CF testing is included as part of the newborn screen in all states.8 Newborn screening (NBS) for CF can include testing for immunoreactive trypsinogen (IRT), which is a pancreatic enzyme found at elevated levels in the blood of some individuals with CF, genetic testing for common CF mutations, or a combination of IRT and genetic testing. Positive newborn screens are typically followed by genetic testing (if not already done), and a sweat test, which measures the chloride concentration in sweat. The sweat test historically has been considered the “gold standard” for CF diagnosis. The presence of two disease-causing mutations also may be considered diagnostic of the disease, even in the absence of classic symptoms or in the case of a negative or inconclusive sweat test.9
In addition to NBS, CF carrier screening (PCS) has become commonplace in the U.S., particularly among pregnant individuals and couples planning a pregnancy. PCS has been found to markedly reduce CF birth rates with a shift towards milder mutations, but it was often avoided for cultural reasons necessitating the use of complementary PCS and NBS.10
A subset of 23 mutations accounts for most cystic fibrosis cases in the U.S. and were accepted by most guidelines as the primary genes to be screened for diagnosis of CF and carrier status. The most common CF-causing mutation is F508del, which is present in over 70% of known CF cases. The Clinical and Functional Translation of CFTR (CFTR2) project continually evaluates genotype and phenotype correlations and has confirmed many additional mutations as being causative of CF.6 Information from over 88,000 patients with specific cystic fibrosis variants from the United States, Canada, and Europe was collected by the CFTR2 team from national CF Patient Registries and placed in the CFTR2 database and then compiled into the CFTR2 website that contains information about the 322 most common CFTR variants.6
Analysis of data from the CF Foundation Patient Registry found that patients of Hispanic, black, or Asian ancestry were less likely to have two identified CFTR variants and more likely to carry no mutations on the commonly used 23 mutation carrier screening panel.11 Analysis of the Exome Aggregation Consortium dataset also found that none of the current genetic screening panels or existing CFTR mutation databases covered most deleterious variants in any geographical population outside of Europe. Both clinical annotation and mutation coverage by commercially available targeted screening panels for CF are strongly biased toward detection of reproductive risk in persons of European descent.12 This research indicates the possible need for adjustment of this panel to facilitate equity in mutation detection between white and nonwhite or mixed-ethnicity CF patients, enabling an earlier diagnosis improving their quality of life.
Proprietary Testing
There are several traditional cystic fibrosis genotyping assays approved by the FDA. eSensor Cystic Fibrosis Carrier Detection System detects 24 CFTR variants,13 Tag-It™ Cystic Fibrosis Kit detects 39 variants,14 Cystic Fibrosis Genotyping Assay detects 30 variants,15 InPlex™ CF Molecular Test detects 32 variants,16 Verigene®CFTR and Verigene®CFTR PolyT Nucleic Acid Tests detect 26 variants,17 Diasorin xTAG® Cystic Fibrosis (CFTR) 39 kit v2 detects 39 variants and the Diasorin xTAG® Cystic Fibrosis (CFTR) 60 kit v2 detects an additional 21 variants,18 and Illumina MiSeqDx™ Cystic Fibrosis 139-Variant Assay detects 139 variants.19These panels target common mutations prevalent in certain populations, providing rapid and cost-effective results. However, they may miss rare or novel mutations not included in the panel, potentially leading to incomplete genetic assessments.
Next generation sequencing (NGS) offers a comprehensive analysis by sequencing the entire CFTR gene enabling the detection of both common and rare mutations, including novel variants that traditional panels might overlook. FDA-approved platforms, such as the Illumina MiSeqDx Cystic Fibrosis Clinical Sequencing Assay, facilitate the detection of both common and rare CFTR mutations.20 A study using NGS for NBS found that the NGS assay was 100% concordant with traditional methods. Retrospective analysis results indicate an IRT/NGS screening algorithm would enable high sensitivity, better specificity, and positive predictive value.21 Further studies have shown that implementing NGS has improved diagnostic accuracy and increased the positive predictive value in newborn screening.22 Additionally, whole genome sequencing of the CFTR gene is also growing in popularity. This type of sequencing has revealed a high prevalence of the intronic variant c.3874-4522A>G in those affected by CF.23
Cystic fibrosis transmembrane conductance regulator (CFTR) modulators are a new class of medications targeting the underlying defect in CF by improving production, intracellular processing, and/or function of the defective CFTR protein. Ivacaftor (IVA), which improves chloride channel function, IVA combined with lumacaftor (LUM), which partially corrects the CFTR misfolding, and IVA combined with tezacaftor, which improves the intracellular processing and trafficking of CFTR, have been approved by the U.S. Food and Drug Administration for use in patients with CF. Additional approval of Trikafta, a combination therapy of elexacaftor, tezacaftor, and ivacaftor, has expanded treatment options by demonstrating efficacy in patients with a broader spectrum of CFTR mutations.24 The indications and efficacy of these drugs depend upon the CFTR mutation in the individual patient. For example, ivacaftor has been FDA-approved and Clinical Pharmacogenetics Implementation Consortium (CPIC)-recommended only for CF patients with the G551D mutation. It is not recommended for any other CF mutation.25
Analytical Validity
Lyon, et al. (2014) assessed clinical laboratories’ proficiency at evaluating genetic alterations for CF. A total of 357 labs participated, performing approximately 120,000 tests monthly. Analytical sensitivity and specificity were 98.8% and 99.6%, respectively. Clinical interpretation matched intended response for zero, one, and two mutations. The authors concluded that laboratory testing for CF in the United States is of high quality.26
Sugunaraj, et al. (2019) completed a cross-sectional study which included the analysis of CFTR variants in 50,778 exomes. Only 24 patients were identified to contain bi-allelic pathogenic CFTR variants; the authors identified 21 of these cases as true-positives and three as potential false positives. Therefore, “genomic screening exhibited a positive predictive value of 87.5%, negative predictive value of 99.9%, sensitivity of 95.5%, and a specificity of 99.9%.”27 Overall, the presence and/or absence of CFTR variants was strongly related to a CF diagnosis.
Hendrix, et al. (2020) assessed the performance of dried blood spot DNA extraction methods in NGS for cystic fibrosis. A total of 20 DNA samples were used in this study and sequenced using Illumina's MiSeqDx™ Cystic Fibrosis 139-Variant Assay and Swift Biosciences’ Accel-Amplicon™ CFTR panel. The MiSeqDx CFTR Clinical Sequencing Assay identified 37 CF-causing variants in the 20 samples tested, for a total of 141 benign variants which were all confirmed with Sanger sequencing. The Swift Biosciences’ Accel-Amplicon™ CFTR panel identified 39 CF-causing variants for a total of 123 benign variants which were all confirmed with Sanger sequencing. Illumina's MiSeqDx performed well with a very high average coverage per method (>3000×) despite most DNA extraction methods being low-concentration and relatively crude. Because of the higher input of samples on a single flow cell, the average coverage per target was lower in the Accel-Amplicon™ CFTR panel in comparison to the clinical sequencing assay, but still often had >1000× coverage. The authors conclude that “both these tests are appropriate DNA extraction methods that can be used with dried blood spots that will result in consistent, high-quality sequencing results.”28
Sicko, et al. (2021) validated the use of a custom NGS assay, Archer CF assay, for CF newborn screening. According to the New York State, CF NBS had high false positive rates. However, after the implementation of this three-tier IRT-DNA-SEQ approach using commercially available tests, the positive predictive value of CF NBS improved from 3.7% to 25.2%. The Archer CF assay involves full gene sequencing, analyzing a specific panel of 338 clinically relevant CFTR variants (second tier), followed by unblinding of all sequence variants and bioinformatic assessment of deletions/duplications (third tier). 227 infant samples were taken, and 190 of 227 carries one or more of the 338 panel variants. The second-tier panel variants were all called correctly, detecting 44 different variants with a 100% sensitivity and 100% specificity. All 227 samples also underwent third tier analysis which detected 62 additional unique SNV/indel variants.22
Pedersen, et al. (2024) evaluated the 11-year performance of the cystic fibrosis newborn bloodspot screening (CF-NBS) and found strong sensitivity (96%) but moderate positive predictive value (PPV) (25%). The program uses an immunoreactive trypsinogen (IRT)-DNA algorithm with a second IRT (IRT-2) as a safety net. Optimal cut-offs for IRT-1 and IRT-2 were identified at 2.7 and 5.9 z-scores, respectively. Simulated algorithm modifications suggested that removing the safety net could increase PPV to 30%, reduce unnecessary second heel-prick tests, and maintain sensitivity at 95%. The findings highlight the potential for optimizing CF-NBS algorithms to improve PPV and reduce follow-up testing while maintaining high diagnostic sensitivity.29
Clinical Utility and Validity
Sosnay, et al. (2013) performed a comprehensive analysis of CF genotypes and phenotypes. The authors collected genotype and phenotype data from 39696 CF patients and found 159 variants with over 0.01% allele frequency, 127 of which met both the clinical and functional criteria for CF. The phenotypic criterion used to define the pathogenic threshold was sweat chloride conductance of 10%. These 127 pathogenic genetic variants were estimated to represent 95.4% of CF alleles, leaving only 0.21% of CF patients without an identified pathological CFTR variant. The phenotypes of these variants vary.30
Wainwright, et al. (2015) evaluated the combination of IVA and LUM in CF patients homozygous for the F508D mutation (labeled Phe508del). A total of 1108 patients were examined. The mean baseline forced expiratory volume in one second “FEV1” was 61% of the predicted value. The absolute mean improvement in FEV1% was found to be 2.6-4%, which corresponded to a 4.3-6.7% relative treatment difference. The rate of pulmonary exacerbations was found to be 30-39% less in the treatment group compared to the placebo group. However, the rate of discontinuation due to an adverse event was 4.2% in the treatment group compared to the placebo group.31
Sharma, et al. (2018) performed a cost-effectiveness analysis of the FDA-approved IVA-LUM combination for the F508D mutation. The authors built a Markov-state model to assess the IVA-LUM for 12-year-old CF patients over several periods of time. “Markov states included mild CF (percentage of predicted forced expiratory volume in one second or FEV1 > 70%), moderate (FEV1 40–70%), severe (FEV1 < 40%) disease, post-transplant, and death. Pulmonary exacerbation and lung transplant were included as transition states.” The IVA-LUM combination resulted in higher quality adjusted life years (QALY, 7.29 vs 6.84 for usual care) but at a cost of $1,778,920.88 per QALY compared to $116,155.76 for usual care. Both monetary amounts were over a 10-year period. The IVA-LUM combination was cost-effective at a threshold of $150,000 / QALY, which was estimated to occur at an annual drug cost of $4153.32
Kessels, et al. (2020) completed a systematic review of prenatal genetic testing for CF. Eight different databases were researched for this review. The authors found that after genetic testing “A change in clinical management was observed: termination of pregnancy (TOP) occurred in most cases where two pathogenic variants were identified in a fetus of carrier parents (158/167; 94.6%).”33 The authors conclude by stating that genetic testing for CF had good diagnostic performance and lead to fewer births affected by CF.
Avram, et al. (2021) studied the cost-effectiveness of genotyping versus sequencing for prenatal CF carrier screening using three strategies: genotyping (“genotyping included the standard 23-variant panel recommended by ACMG/ACOG”) both partners, genotyping one partner then sequencing the second, and sequencing both partners. The authors used a decision-analytic model to generate a theoretical cohort of four million pregnant people and their partners. Sequencing both partners identified 1,099 carrier couples that would have been missed by genotyping of both partners, resulting in 273 fewer missed prenatal diagnoses, 152 more terminations, 152 fewer affected newborns, and screening with genotyping followed by sequencing identified 477 more carrier couples as compared to genotyping both partners, resulting in 119 fewer missed prenatal diagnoses, 66 more terminations, and 66 fewer affected newborns. The incremental cost-effectiveness ratio of genotyping followed by sequencing compared to genotyping both partners was $180,004/ quality-adjusted life year (QALY) and the incremental cost-effectiveness ratio of sequencing both partners compared to genotyping followed by sequencing was $17.6 million/QALY. “Compared to genotyping both partners, the cost per CF case averted for genotyping/sequencing was $1.2 million and sequencing both partners was $49.4 million. . . Sequencing both partners was cost-effective when the cost of sequencing fell below $339 per test. Genotyping/sequencing was cost-effective when the cost of sequencing was between $340 and 1837 per test. Lastly, genotyping both partners was cost-effective when sequencing cost above $1838 per test". The authors concluded that “before being routinely recommended, sequencing for CF carrier screening requires greater examination at a population-based level.34
American College of Medical Genetics and Genomics (ACMG)
The ACMG recommends offering CF carrier screening to all individuals capable of becoming pregnant who are of reproductive age, regardless of ethnic background. Ideally, the testing would occur prior to pregnancy, but if the individual is pregnant, then they should be tested as early as possible during the pregnancy. ACMG also indicates that if the individual is found to be a CF carrier, then their reproductive partner should also be tested.35 If the individual presents for testing late in the pregnancy, simultaneous testing of both reproductive partners of the fetus should be considered. When there is a paternal family history of CF, carrier testing of the father is warranted.
In 2020, the ACMG introduced technical guidelines for CFTR variant testing, particularly emphasizing considerations during pregnancy. These guidelines suggest that simultaneous testing might be necessary based on various factors like gestational age, family history, ethnicity, or patient preferences. Carrier testing is recommended for individuals with a family history of CF, partners of those with such history, partners of males with CAVD, reproductive age women, and gamete donors. Additionally, CFTR variant testing can be conducted for prenatal diagnosis using cells obtained through procedures like amniocentesis or chorionic villus sampling (CVS). To ensure inclusivity, “the ACMG recommends either a classification-based reporting approach or a classification-based (targeted) testing approach (which has historically been used for CFTR carrier screening). For those laboratories who wish to continue using a targeted testing approach, the ACMG-23 variant panel remains as the minimum list of CFTR variants that should be included. Laboratories may want to consider adding additional variants to their panel depending on the ethnic composition of their expected test population. However, the minimum list of CFTR variants recommended for pan-ethnic carrier screening has not been increased at this time.”36
In 2023, the ACMG issued updated recommendations for CFTR carrier screening, introducing a new minimum CFTR variant set (Table 1) that replaces the previous set of 23 variants. These revised recommendations specifically apply to carrier screening and do not affect CFTR variant testing for diagnosis or newborn screening. However, all other aspects of the 2020 ACMG CFTR technical standards remain unchanged.37
Table 1 (adapted from Deignan, et al. (2023)) : CFTR Carrier Screening Variant Set (n=100)
| DNA Variant |
Protein Variant |
Legacy Name |
| c.4C>T |
p.Gln2Ter |
Q2X |
| c.178G>T |
p.Glu60Ter |
E60X |
| c.200C>T |
p.Pro67Leu |
P67L |
| c.223C>T |
p.Arg75Ter |
R75X |
| c.254G>A |
p.Gly85Glu |
G85Ea |
| c.262_263del |
p.Leu88IlefsTer22 |
394delTT |
| c.271G>A |
p.Gly91Arg |
G91R |
| c.274-1G>A |
p.? |
406-1G->A |
| c.292C>T |
p.Gln98Ter |
Q98X |
| c.293A>G |
p.Gln98Arg |
Q98R |
| c.313del |
p.Ile105SerfsTer2 |
444delA |
| c.328G>C |
p.Asp110His |
D110H |
| c.349C>T |
p.Arg117Cys |
R117C |
| c.350G>A |
p.Arg117His |
R117Ha |
| c.489+1G>T |
p.? |
621+1G->Ta |
| c.571T>G |
p.Phe191Val |
F191V |
| c.579+1G>T |
p.? |
711+1G->Ta |
| c.579+3A>G |
p.? |
711+3A->G |
| c.617T>G |
p.Leu206Trp |
L206W |
| c.653T>A |
p.Leu218Ter |
L218X |
| c.695T>A |
p.Val232Asp |
V232D |
| c.803del |
p.Asn268IlefsTer17 |
935delA |
| c.868C>T |
p.Gln290Ter |
Q290X |
| c.988G>T |
p.Gly330Ter |
G330X |
| c.1000C>T |
p.Arg334Trp |
R334Wa |
| c.1013C>T |
p.Thr338Ile |
T338I |
| c.1021_1022dup |
p.Phe342HisfsTer28 |
1154insTC |
| c.1029del |
p.Cys343Ter |
1161delC |
| c.1040G>A |
p.Arg347His |
R347H |
| c.1040G>C |
p.Arg347Pro |
R347Pa |
| c.1055G>A |
p.Arg352Gln |
R352Q |
| c.1155_1156dup |
p.Asn386IlefsTer3 |
1288insTA |
| c.1327_1330dup |
p.Ile444ArgfsTer3 |
1461ins4 |
| c.1364C>A |
p.Ala455Glu |
A455Ea |
| c.1367T>C |
p.Val456Ala |
V456A |
| c.1373del |
p.Gly458AspfsTer11 |
1504delG |
| c.1393-1G>A |
p.? |
1525-1G->A |
| c.1397C>G |
p.Ser466Ter |
S466X |
| c.1400T>C |
p.Leu467Pro |
L467P |
| c.1519_1521del |
p.Ile507del |
I507dela |
| c.1521_1523del |
p.Phe508del |
F508dela |
| c.1572C>A |
p.Cys524Ter |
C524X |
| c.1584+1G>A |
p.? |
1716+1G->A |
| c.1585-1G>A |
p.? |
1717-1G->Aa |
| c.1624G>T |
p.Gly542Ter |
G542Xa |
| c.1646G>A |
p.Ser549Asn |
S549N |
| c.1647T>G |
p.Ser549Arg |
S549R |
| c.1651G>A |
p.Gly551Ser |
G551S |
| c.1652G>A |
p.Gly551Asp |
G551Da |
| c.1657C>T |
p.Arg553Ter |
R553Xa |
| c.1673T>C |
p.Leu558Ser |
L558S |
| c.1675G>A |
p.Ala559Thr |
A559T |
| c.1679G>C |
p.Arg560Thr |
R560Ta |
| c.1679+1G>A |
p.? |
1811+1G->A |
| c.1680-886A>G |
p.? |
1811+1634A->G |
| c.1680A>C |
p.Arg560Ser |
R560S |
| c.1682C>A |
p.Ala561Glu |
A561E |
| c.1692del |
p.Asp565MetfsTer7 |
1824delA |
| c.1705T>G |
p.Tyr569Asp |
Y569D |
| c.1753G>T |
p.Glu585Ter |
E585X |
| c.1766+1G>A |
p.? |
1898+1G->Aa |
| c.1766+5G>T |
p.? |
1898+5G->T |
| c.1837G>A |
p.Ala613Thr |
A613T |
| c.1882G>A |
p.Gly628Arg |
G628R |
| c.2052dup |
p.Gln685ThrfsTer4 |
2184insA |
| c.2052del |
p.Lys684AsnfsTer38 |
2184delAa |
| c.2125C>T |
p.Arg709Ter |
R709X |
| c.2175dup |
p.Glu726ArgfsTer4 |
2307insA |
| c.2290C>T |
p.Arg764Ter |
R764X |
| c.2353C>T |
p.Arg785Ter |
R785X |
| c.2374C>T |
p.Arg792Ter |
R792X |
| c.2490+1G>A |
p.? |
2622+1G->A |
| c.2657+5G>A |
p.? |
2789+5G->Aa |
| c.2668C>T |
p.Gln890Ter |
Q890X |
| c.2739T>A |
p.Tyr913Ter |
Y913X |
| c.2834C>T |
p.Ser945Leu |
S945L |
| c.2909G>A |
p.Gly970Asp |
G970D |
| c.2988G>A |
p.Gln996= |
3120G->A |
| c.2988+1G>A |
p.? |
3120+1G->Aa |
| c.3067_3072del |
p.Ile1023_Val1024del |
3199del6 |
| c.3107C>A |
p.Thr1036Asn |
T1036N |
| c.3140-26A>G |
p.? |
3272-26A->G |
| c.3196C>T |
p.Arg1066Cys |
R1066C |
| c.3197G>A |
p.Arg1066His |
R1066H |
| c.3266G>A |
p.Trp1089Ter |
W1089X |
| c.3294G>C |
p.Trp1098Cys |
W1098C |
| c.3353C>T |
p.Ser1118Phe |
S1118F |
| c.3472C>T |
p.Arg1158Ter |
R1158X |
| c.3484C>T |
p.Arg1162Ter |
R1162Xa |
| c.3528del |
p.Lys1177SerfsTer15 |
3659delCa |
| c.3612G>A |
p.Trp1204Ter |
W1204X |
| c.3659del |
p.Thr1220LysfsTer8 |
3791delC |
| c.3717+5G>A |
p.? |
3849+5G->A |
| c.3718-2477C>T |
p.? |
3849+10kbC->Ta |
| c.3744del |
p.Lys1250ArgfsTer9 |
3876delA |
| c.3764C>A |
p.Ser1255Ter |
S1255X |
| c.3808del |
p.Asp1270MetfsTer8 |
3940delG |
| c.3846G>A |
p.Trp1282Ter |
W1282Xa |
| c.3889dup |
p.Ser1297PhefsTer5 |
4016insT |
| c.3909C>G |
p.Asn1303Lys |
N1303Ka |
aVariants that were part of the previously recommended minimum 23-variant set.
The ACMG also recommends that testing for the IVS8 polyT variant be performed only when carrier screening reveals a R117H mutation, as the polyT variant influences the clinical severity of the mutation but does not cause CF by itself.38
The ACMG provides guidance on indications for CFTR variant testing. Diagnostic testing can be used for the molecular confirmation of a clinical CF diagnosis, for infants with meconium ileus, for individuals with CAVD, individuals with idiopathic pancreatitis or bronchiectasis, and as a follow-up to newborn screening. Regarding carrier testing, ACMG recommends that carrier testing be offered to individuals with a positive family history of CF, to partners of individuals with a positive family history, to partners of individuals with CAVD, to reproductive age individuals capable of becoming pregnant, and to gamete donors.36
Prenatal testing can be performed using cells obtained for diagnostic cytogenetic testing (i.e., amniocentesis or chorionic villus sampling [CVS]). Testing can take place on cultured or uncultured amniocytes or villous trophoblasts. According to the guidelines, targeted sequencing for specific CFTR variants may be considered when:
- “A pathogenic or likely pathogenic variant is confirmed in both partners.
- A pathogenic or likely pathogenic variant is confirmed in one partner and a VUS or variant associated with variable expressivity is confirmed in the other partner
- As part of preimplantation genetic testing when both biological parents are confirmed carriers of a pathogenic or likely pathogenic variant.”36
Comprehensive CFTR sequencing may be considered when:
- One member of a couple is known to be a carrier of a pathogenic or likely pathogenic variant and any of the following is also true:
- The partner is unavailable for screening
- The partner has not been screened and any of the following is also true:
- o Screening that partner would be cost prohibitive
- o The results from the partner would not be available in time to allow for reproductive decision making
- o A diagnostic procedure (e.g., CVS, amniocentesis) is also being performed for other reasons (e.g., ultrasound abnormality).
- An ultrasound finding (i.e., fetal echogenic bowel) suggests an affected fetus and CFTR variant information is not available from either biological parent.”36
The ACMG recommends the following regarding testing of intronic variants, deletions/duplications, reporting VUS and variable expressivity, and reporting of intron 9 polyT and TG regions:
- “ACMG recommends the reporting of the c.2657+5G>A and c.3718–2477C>T intronic variants. If these variants are not detectable with the laboratory’s methodology (e.g., exome sequencing), then a separate assay should be performed to test for these variants.
- For all prenatal, postnatal, and adult diagnostic testing and carrier screening indications for CFTR testing, the ACMG does not recommend the testing of any specific exon-level or gene-level deletion or duplication variants.
- For all prenatal, postnatal, and adult diagnostic testing indications for CFTR where comprehensive methods are used, the ACMG recommends the reporting of VUS. For all adult carrier screening indications for CFTR where comprehensive methods are used, VUS should generally not be reported. However, laboratories may want to consider reporting VUS in the partner of an individual who had a pathogenic or likely pathogenic variant detected during screening.
- For all prenatal, postnatal, and adult diagnostic testing indications for CFTR where comprehensive methods are used, the ACMG recommends the reporting of any variants associated with variable expressivity.
- For all prenatal, postnatal, and adult diagnostic testing indications for CFTR, the ACMG recommends the reporting of R117H status as well as the results from at least the associated polyT tract. For all adult carrier screening indications for CFTR, polyT status should be reported when the R117H variant is detected; laboratories may also want to consider reporting the results from the associated polyT tract in the partner of an individual who had a pathogenic or likely pathogenic variant detected during screening.”36
The ACMG released recommendations for preconception and prenatal carrier screening. The following tier-based approach to carrier screening was produced and recommendations were made:39
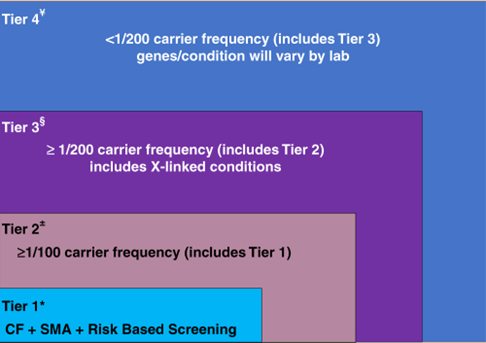
CF: cystic fibrosis, SMA: spinal muscular atrophy.
- Preconception screening is recommended over prenatal screening.
- Concurrent partner testing should be offered if testing done during pregnancy.
- Tier 3 screening is recommended which includes 97 autosomal recessive genes and 16 X-linked genes, including DMD and Fragile X.
- The second biological parent may be offered Tier 3 carrier screening (for autosomal recessive conditions) when carrier screening is being performed at the same time in their reproductive partner who is capable of becoming pregnant.
- All pregnant patients and those planning a pregnancy should be offered Tier 3 carrier screening.
- Tier 4 screening should be considered when a pregnancy arises from a known or possible consanguineous relationship (second cousins or closer) or when a family or personal medical history warrants further risk assessment.
- ACMG does not recommend offering Tier 1 and/or Tier 2 screening, because these do not provide equitable evaluation of all racial/ethnic groups and routine offering of Tier 4 panels.39
Cystic Fibrosis Foundation
The Cystic Fibrosis Foundation convened a group of to develop clear and actionable consensus guidelines on the diagnosis of CF and to clarify diagnostic criteria and terminology for other disorders associated with CFTR mutations. The experts determined that “diagnoses associated with CFTR mutations in all individuals, from newborn to adult, be established by evaluation of CFTR function with a sweat chloride test. The latest mutation classifications annotated in the Clinical and Functional Translation of CFTR project (http://www.cftr2.org/index.php) should be used to aid in diagnosis.”40
The committee approved 27 consensus statements:40
| 1 |
Sweat chloride testing should be performed according to approved procedural guidelines published in established, international protocols such as the CLSI 2009 Guidelines. |
|
| 2 |
Newborns with a positive CF newborn screen, to increase the likelihood of collecting an adequate sweat specimen, should have the test performed bilaterally and when the infant weighs >2 kg, and is at least 36 wk of corrected gestational age. |
|
| 3 |
Newborns greater than 36 wk gestation and >2 kg body weight with a positive CF newborn screen, or positive prenatal genetic test, should have sweat chloride testing performed as soon as possible after 10 d of age, ideally by the end of the neonatal period (4 wk of age). |
|
| 4 |
In infants with presumptive CF identified through NBS, CF treatment should not be delayed while efforts to establish a diagnosis of CF are initiated. |
|
| 5 |
Sweat chloride analysis should be performed within a few hours of sweat collection and the results and interpretations should be reported to clinicians and parents or patients, as soon as possible and certainly on the same day. |
|
| 6 |
In individuals presenting with a positive newborn screen, clinical features consistent with CF, or a positive family history, a diagnosis of CF can be made if the sweat chloride value is ≥60 mmol/L. |
|
| 7 |
Individuals who are screen-positive and meet sweat chloride criteria for CF diagnosis should undergo CFTR genetic testing if the CFTR genotype was not available through the screening process or is incomplete. |
|
| 8 |
In individuals with a positive newborn screen, a sweat chloride <30 mmol/L indicates that CF is unlikely. |
|
| 9 |
Individuals with clinical features that may be consistent with CF who have a sweat chloride <30 mmol/L indicates that CF is less likely. It may, however, be considered if evolving clinical criteria and/or CFTR genotyping support CF and not an alternative diagnosis. |
|
| 10 |
Individuals presenting with a positive newborn screen, symptoms of CF, or a positive family history, and sweat chloride values in the intermediate range (30-59 mmol/L) on two separate occasions may have CF. They should be considered for extended CFTR gene analysis and/or CFTR functional analysis. |
|
| 11 |
The latest classifications identified in the CFTR2 project (http://www.cftr2.org/index.php) should be used to aid with CF diagnosis: Mutation of varying clinical consequence (MVCC): a mutation that in combination with a CF-causing mutation or another MVCC mutation may result in CF Uncharacterized mutation/mutation of UNK: mutation that has not been evaluated by CFTR2 and may be disease causing or of variable clinical consequence or benign Non-CF-causing mutation: individuals with 1 or more are unlikely to have CF (as a result of that allele) |
|
| 12 |
In individuals presenting with a positive newborn screen, symptoms of CF, or a positive family history, the identification of 2 CF-causing mutations (defined by CFTR2) is consistent with a diagnosis of CF. Sweat chloride testing is necessary, though, to confirm the diagnosis. |
|
| 13 |
The absence of detection of 2 CF-causing CFTR mutations does not exclude a diagnosis of CF. |
|
| 14 |
If further CF functional testing is needed (NPD and ICM), it should be performed in a validated reference center with trained staff certified by the CF Foundation TDN or ECFS Clinical Trial Network. |
|
| 15 |
In individuals with a positive newborn screen but variable or uncharacterized CFTR mutations (<2 CF-causing mutations), the diagnosis of CF can be made by demonstrating CFTR dysfunction (a sweat chloride ≥ 60 mmol/L or CF-typical NPD or ICM). |
|
| 16 |
The term CRMS [CFTR-related metabolic disorder] is used in the US for healthcare delivery purposes and CFSPID [cystic fibrosis screen positive, inconclusive diagnosis] is used in other countries, but these both describe an inconclusive diagnosis following NBS. |
|
| 17 |
The term CRMS/CFSPID is reserved for individuals who screen positive without clinical features consistent with a diagnosis of CF. |
|
| 18 |
The definition of CRMS/CFSPID is an infant with a positive NBS test for CF and either: OR An intermediate sweat chloride value (30-59 mmol/L) and 1 or 0 CF-causing mutations |
|
| 19 |
Children designated as CRMS/CFSPID should undergo at least one repeat sweat chloride test at CF centers with suitable expertise, such as an accredited CF center. |
|
| 20 |
Children designated as CRMS/CFSPID should have clinical evaluation performed by CF providers to identify the minority that may develop clinical symptoms. |
|
| 21 |
Children designated as CRMS/CFSPID can be considered for extended CFTR gene analysis (sequencing and or deletion duplication testing), as well as CFTR functional analysis (NPD/ICM) testing to further define their likelihood of developing CF. |
|
| 22 |
The decision to reclassify children designated as CRMS/CFSPID as CF is an integrated decision that should take into account functional assessment of CFTR (sweat chloride, and possibly NPD/ICM), CFTR genetic analysis, and clinical assessment by the CF clinicians caring for the patient. |
|
| 23 |
Genetic counseling should be offered to families of individuals followed for CRMS/CFSPID, including a discussion of the risk in future pregnancies. |
|
| 24 |
Research Recommendation: Infants with a designation of CRMS/CFSPID (by definition) do not have clinical features consistent with a diagnosis of CF and further research is needed to determine the prognosis and best practices for frequency and duration of follow-up. |
|
| 25 |
For individuals presenting with CF symptoms, the same diagnostic criteria recommended for the screened population for sweat chloride testing, CFTR genetic analysis, and CFTR functional testing should be used to confirm a CF diagnosis. |
|
| 26 |
The diagnosis of CFTR-related disorder has been defined as a monosymptomatic clinical entity (CBAVD/pancreatitis/bronchiectasis) associated with CFTR dysfunction that does not fulfill the diagnostic criteria for CF. |
|
| 27 |
Clinicians should avoid the use of terms like classic/nonclassic CF, typical/atypical CF, delayed CF, because these terms have no harmonized definition and could be confusing for families or caregivers. |
|
The Cystic Fibrosis Foundation also convened a multidisciplinary committee of CF caregivers to develop evidence-based guidelines for CFTR modulator therapy. The committee defined patients with CF as individuals who met the above CFF criteria for diagnosis of CF, combined with evidence of abnormal CFTR function, as demonstrated by elevated sweat chloride, detection of two CF-causing CFTR mutations, or abnormal nasal potential differences.41
For adults and children aged six years and older with CF due to gating mutations other than G551D or R117H, the guideline panel made a conditional recommendation for treatment with IVA. For those with the R117H mutation, the guideline panel made a conditional recommendation for treatment with IVA for 1) adults aged 18 years or older, and 2) children aged 6–17 years with a forced expiratory volume in one second (FEV1) less than 90% predicted. For those with the R117H mutation, the guideline panel made a conditional recommendation against treatment with IVA for 1) children aged 12–17 years with an FEV1 greater than 90% predicted, and 2) children less than six years of age. Among those with two copies of F508del, the guideline panel made a strong recommendation for treatment with IVA/LUM for adults and children aged 12 years and older with an FEV1 less than 90% predicted; and made a conditional recommendation for treatment with IVA/LUM for 1) adults and children aged 12 years or older with an FEV1 greater than 90% predicted, and 2) children aged 6–11 years.41
In an updated 2024 report, a multidisciplinary committee with the Cystic Fibrosis Foundation released additional recommendations as described below:
Genetic Testing:
- “The CFF recommends that people with CRMS/CFSPID who have <2 disease-causing variants identified by NBS should undergo sequencing of the coding and flanking regions and deletion/duplication (del/dup) analysis of the coding and exon flanking regions of CFTR (Grade B).
- The CFF recommends, for people with CRMS/CFSPID, selectively offering full gene CFTR sequencing including intronic regions when the CFTR genotype remains incomplete after coding and flanking region sequencing and del/dup (Grade C)
- The CFF recommends CFTR genetic evaluation for parents of people with CRMS/CFSPID when phasing the CFTR variants (ie, in cis or trans) would inform the diagnostic status of the individual by confirming the inheritance pattern (Grade A).
- The CFF recommends offering CFTR genetic evaluation for siblings of people with CRMS/CFSPID (Grade B).
- The CFF recommends, for families of people with CRMS/CFSPID, that health care professionals (HCPs) providing genetic counseling should have training or clinical expertise in CF and genetics. A licensed or certified genetic counselor (GC) should be accessible to families of people with CRMS/CFSPID for further support, including discussions regarding future reproductive decision-making (Grade B).”42
Monitoring CFTR diagnosis:
- “The CFF recommends, for people with CRMS/CFSPID, at least annual follow-up by a CF clinician and nurse, with an initial assessment to include a social worker, a mental health coordinator (MHC), and/or a genetic counseling provider. Continued follow-up by a social worker, MHC, and/or genetic counselor should be part of the care of CRMS/CFSPID, depending on the needs of that individual and family (Grade B)
- The CFF recommends for people with CRMS/CFSPID to repeat SCT at 6 months of life and annually, at least until age 8 years (Grade B).”42
American College of Obstetrics and Gynecology (ACOG)
The ACOG recommends offering CF carrier screening to all individuals who are considering becoming pregnant or who are currently pregnant. Expanded mutation panels which enhance the sensitivity for carrier screening can be considered for carrier detection, especially in non-Caucasian ethnic groups, but testing should not be repeated if performed previously (e.g., during previous pregnancy). If the patient is found to be a carrier, then her partner should be tested. They also indicate that newborn screening is not a replacement for carrier screening in a population.
Prenatal diagnosis is indicated after genetic counseling if both parents are carriers, or if the mother is a carrier and the father is unknown or unavailable for testing.
The ACOG recommends that complete gene sequencing of the CTFR gene is not appropriate for use in carrier screening, rather reserved for patients with cystic fibrosis, patients with negative carrier screening but a family history of cystic fibrosis, individuals with congenital bilateral absence of the vas deferens, or newborns with a positive newborn screening after the standard 23 gene screen has a negative result.
They also recommend referral to genetic counseling for couples in which both partners are CF carriers, prenatal diagnosis, and advanced reproductive technologies to decrease the risk of affected offspring should be discussed.43
These statements were reaffirmed in 2023.
National Society of Genetic Counselors (NSGC)
The National Society of Genetic Counselors (NSGC) recommends that for all individuals capable of becoming pregnant and who are of reproductive age, carrier screening for CG should be offered “regardless of ancestry; preferably pre-conceptionally. CF carrier testing should also be offered to any individual with a family history of CF and to partners of mutation carriers and people with CF.”44\
Regarding what mutations should be included in the carrier screening test, the NSGC states, “Carrier testing panels should include the mutations recommended by ACOG and ACMG. For individuals of non- Northern European descent, pan-ethnic panels that include additional mutations more commonly identified in minority populations are appropriate to consider. Focus general population CF screening practices on identifying carriers of established disease-causing CFTR mutations.”44
The NSGC agrees with the ACMG regarding testing for IVS 8 5T/7T/9T as a reflex when mutation R117H is found in the CF carrier screen. They also assert that “in the absence of an R117H mutation, assessment of the intron 8 polyT or TG tracts is not recommended for routine CF carrier testing.”44
European Cystic Fibrosis Society (ECFS)
The ECFS mentioned CFTR modulators in their standards of care guideline. Recommendations state that “in patients with the G551D mutation ivacaftor should be part of standard of care”; at the time of writing, only one CFTR modulator had been shown to “demonstrate clinical efficacy.”45
In 2018, the ECFS published a revised version of their best practice guidelines. The ECFS has published the following requirement to when providing a CF diagnosis:
- “To be able to perform genetic testing for the most appropriate panel of CFTR mutations for the local population. Access to extended exon DNA analysis should be available when required” 46.
The ECFS also states that genetic counseling should be offered when reporting a CF diagnosis to a patient.
National Institute for Health and Care Excellence (NICE)
In 2017, the NICE published guidelines on the diagnosis and management of CF. NICE includes the following recommendations on CF diagnoses:
- “Be aware that cystic fibrosis can be diagnosed based on:
- positive test results in people with no symptoms, for example infant screening (blood spot immunoreactive trypsin test) followed by sweat and gene tests for confirmation or
- clinical manifestations, supported by sweat or gene test results for confirmation or
- clinical manifestations alone, in the rare case of people with symptoms who have normal sweat or gene test results.
- Assess for cystic fibrosis and, when clinically appropriate, perform a sweat test (for children and young people) or a cystic fibrosis gene test (for adults) in people with any of the following:
- family history
- congenital intestinal atresia
- meconium ileus
- symptoms and signs that suggest distal intestinal obstruction syndrome
- faltering growth (in infants and young children)
- undernutrition
- recurrent and chronic pulmonary disease, such as:
- recurrent lower respiratory tract infections
- clinical or radiological evidence of lung disease (in particular bronchiectasis)
- persistent chest X-ray changes
- chronic wet or productive cough
- chronic sinus disease
- obstructive azoospermia (in young people and adults)
- acute or chronic pancreatitis
- malabsorption
- rectal prolapse (in children)
- pseudo-Bartter syndrome.
- Refer people with suspected cystic fibrosis to a specialist cystic fibrosis center if:
- they have a positive or equivocal sweat test result
- their assessment suggests they have cystic fibrosis but their test results are normal
- gene testing reveals 1 or more cystic fibrosis mutations.”47
These statements were reviewed and reasserted in 2024.
Physicians Committee for Responsible Medicine (PCRM)
The PSRM has published guidelines on diagnosis of cystic fibrosis. Below are the recommended guidelines:
- A sweat chloride test can be performed as early as 48 hours after birth, which is the mainstay of laboratory confirmation.
- Newborn Screening is mandatory in all 50 states.
A patient with symptoms may require a sweat chloride test or genetic testing. “Patients who exhibit signs and symptoms but do not have a positive sweat test should have a genetic test. Prenatal testing and newborn screening may be used. Early detection renders a better clinical course. While not widely available, nasal potential difference measurements can confirm the diagnosis if sweat testing and genetic testing results are inconclusive.”48
Reference
1. Pritchard LL. Respiratory Conditions Update: Cystic Fibrosis. FP essentials. Sep 2016;448:35-43.
2. Cohen-Cymberknoh M, Shoseyov D, Kerem E. Managing cystic fibrosis: strategies that increase life expectancy and improve quality of life. American journal of respiratory and critical care medicine. Jun 1 2011;183(11):1463-71. doi:10.1164/rccm.201009-1478CI
3. Clancy JP, Jain M. Personalized medicine in cystic fibrosis: dawning of a new era. American journal of respiratory and critical care medicine. Oct 1 2012;186(7):593-7. doi:10.1164/rccm.201204-0785PP
4. Barrett PM, Alagely A, Topol EJ. Cystic fibrosis in an era of genomically guided therapy. Human molecular genetics. Oct 15 2012;21(R1):R66-71. doi:10.1093/hmg/dds345
5. Bell SC, De Boeck K, Amaral MD. New pharmacological approaches for cystic fibrosis: promises, progress, pitfalls. Pharmacology & therapeutics. Jan 2015;145:19-34. doi:10.1016/j.pharmthera.2014.06.005
6. CF Foundation U, Johns Hopkins University, The Hospital for Sick Children. The Clinical and Functional TRanslation of CFTR (CFTR2). https://cftr2.org/
7. Wilschanski M, Zielenski J, Markiewicz D, et al. Correlation of sweat chloride concentration with classes of the cystic fibrosis transmembrane conductance regulator gene mutations. The Journal of pediatrics. Nov 1995;127(5):705-10. doi:10.1016/s0022-3476(95)70157-5
8. Grosse SD, Boyle CA, Botkin JR, et al. Newborn screening for cystic fibrosis: evaluation of benefits and risks and recommendations for state newborn screening programs. MMWR Recommendations and reports : Morbidity and mortality weekly report Recommendations and reports. Oct 15 2004;53(Rr-13):1-36.
9. Farrell PM, Rosenstein BJ, White TB, et al. Guidelines for diagnosis of cystic fibrosis in newborns through older adults: Cystic Fibrosis Foundation consensus report. The Journal of pediatrics. Aug 2008;153(2):S4-s14. doi:10.1016/j.jpeds.2008.05.005
10. Stafler P, Mei-Zahav M, Wilschanski M, et al. The impact of a national population carrier screening program on cystic fibrosis birth rate and age at diagnosis: Implications for newborn screening. Journal of cystic fibrosis : official journal of the European Cystic Fibrosis Society. Jul 2016;15(4):460-6. doi:10.1016/j.jcf.2015.08.007
11. Schrijver I, Pique L, Graham S, Pearl M, Cherry A, Kharrazi M. The Spectrum of CFTR Variants in Nonwhite Cystic Fibrosis Patients: Implications for Molecular Diagnostic Testing. The Journal of molecular diagnostics : JMD. Jan 2016;18(1):39-50. doi:10.1016/j.jmoldx.2015.07.005
12. Lim RM, Silver AJ, Silver MJ, et al. Targeted mutation screening panels expose systematic population bias in detection of cystic fibrosis risk. Genetics in medicine : official journal of the American College of Medical Genetics. Feb 2016;18(2):174-9. doi:10.1038/gim.2015.52
13. FDA. eSensor® Cystic Fibrosis Carrier Detection System 510(k) Summary. Updated February 28, 2006. https://pathsurveyor.com/api/pmn_pdf?knumber=K060543
14. FDA. Decision Summary Tag It Cystic Fibrosis Carrier Detection System 510(k) Premarket Notification. https://www.accessdata.fda.gov/cdrh_docs/reviews/K043011.pdf
15. FDA. Decision Summary Cystic Fibrosis Genotyping Assay 510(k) Premarket Notification. https://www.accessdata.fda.gov/cdrh_docs/reviews/K062028.pdf
16. Hologic. InPlex Molecular Test. https://www.hologic.com/sites/default/files/package-insert/Hologic%20CF%20IVD%20Package%20Insert.pdf
17. FDA. Decision Summary Verigene® CFTR and Verigene® CFTR PolyT Nucleic Acid Tests 510(k) Premarket Notification. https://www.accessdata.fda.gov/cdrh_docs/reviews/K083294.pdf
18. Diasorin. xTAG® Cystic Fibrosis (CFTR) 39 kit v2 and 60 kit v2. https://us.diasorin.com/sites/default/files/products-documentation-tool/xTAG%20Cystic%20Fibrosis%20%28CFTR%29%2039%20kit%20v2%20and%2060%20kit%20v2%20%28US-IVD%29.SS50153.0923.APM_.pdf
19. Illumina. Illumina MiSeqDx™ Cystic Fibrosis 139-Variant Assay. https://support.illumina.com/clinical_support/clinical_kits/miseqdx-cf-139-variant-assay.html
20. FDA. Decision Summary Illumina MiSeqDxTM Cystic Fibrosis Clinical Sequencing Assay 510(k) Premarket Notification. https://www.accessdata.fda.gov/cdrh_docs/reviews/K132750.pdf
21. Baker MW, Atkins AE, Cordovado SK, Hendrix M, Earley MC, Farrell PM. Improving newborn screening for cystic fibrosis using next-generation sequencing technology: a technical feasibility study. Genetics in medicine : official journal of the American College of Medical Genetics. Mar 2016;18(3):231-8. doi:10.1038/gim.2014.209
22. Sicko RJ, Stevens CF, Hughes EE, et al. Validation of a Custom Next-Generation Sequencing Assay for Cystic Fibrosis Newborn Screening. Int J Neonatal Screen. Nov 2 2021;7(4)doi:10.3390/ijns7040073
23. Morris-Rosendahl DJ, Edwards M, McDonnell MJ, et al. Whole Gene Sequencing of CFTR Reveals a High Prevalence of the Intronic Variant c.3874-4522A>G in Cystic Fibrosis. American journal of respiratory and critical care medicine. Feb 4 2020;doi:10.1164/rccm.201908-1541LE
24. FDA approves new breakthrough therapy for cystic fibrosis. U.S. Food and Drug Administration; October 21, 2019. https://www.fda.gov/news-events/press-announcements/fda-approves-new-breakthrough-therapy-cystic-fibrosis
25. Clancy JP, Johnson SG, Yee SW, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines for ivacaftor therapy in the context of CFTR genotype. Clinical pharmacology and therapeutics. Jun 2014;95(6):592-7. doi:10.1038/clpt.2014.54
26. Lyon E, Schrijver I, Weck KE, Ferreira-Gonzalez A, Richards CS, Palomaki GE. Molecular genetic testing for cystic fibrosis: laboratory performance on the College of American Pathologists external proficiency surveys. Original Research Article. Genetics In Medicine. 07/31/online 2014;17:219. doi:10.1038/gim.2014.93
27. Sugunaraj JP, Brosius HM, Murray MF, et al. Predictive value of genomic screening: cross-sectional study of cystic fibrosis in 50,788 electronic health records. NPJ Genom Med. 2019;4:21. doi:10.1038/s41525-019-0095-6
28. Hendrix MM, Cuthbert CD, Cordovado SK. Assessing the Performance of Dried-Blood-Spot DNA Extraction Methods in Next Generation Sequencing. International Journal of Neonatal Screening. 2020;6(2):36. doi:10.3390/ijns6020036
29. Pedersen ESL, Cm, Jurca M, et al. Cystic fibrosis newborn screening in Switzerland – evaluation and scenarios for improvement after 11 years of follow-up. Journal of Cystic Fibrosis. 2024;23(4):796-803. doi:10.1016/j.jcf.2024.04.008
30. Sosnay PR, Siklosi KR, Van Goor F, et al. Defining the disease liability of variants in the cystic fibrosis transmembrane conductance regulator gene. Nature genetics. Oct 2013;45(10):1160-7. doi:10.1038/ng.2745
31. Wainwright CE, Elborn JS, Ramsey BW, et al. Lumacaftor-Ivacaftor in Patients with Cystic Fibrosis Homozygous for Phe508del CFTR. The New England journal of medicine. Jul 16 2015;373(3):220-31. doi:10.1056/NEJMoa1409547
32. Sharma D, Xing S, Hung YT, Caskey RN, Dowell ML, Touchette DR. Cost-effectiveness analysis of lumacaftor and ivacaftor combination for the treatment of patients with cystic fibrosis in the United States. Orphanet journal of rare diseases. 2018;13(1):172. doi:10.1186/s13023-018-0914-3
33. Kessels SJM, Carter D, Ellery B, Newton S, Merlin TL. Prenatal genetic testing for cystic fibrosis: a systematic review of clinical effectiveness and an ethics review. Genetics in medicine : official journal of the American College of Medical Genetics. Feb 2020;22(2):258-267. doi:10.1038/s41436-019-0641-8
34. Avram CM, Dyer AL, Shaffer BL, Caughey AB. The cost-effectiveness of genotyping versus sequencing for prenatal cystic fibrosis carrier screening. Prenatal Diagnosis. 2021;41(11):1449-1459. doi:10.1002/pd.6027
35. Pletcher BA, Bocian M. Preconception and prenatal testing of biologic fathers for carrier status. American College of Medical Genetics. Genetics in medicine : official journal of the American College of Medical Genetics. Feb 2006;8(2):134-5. doi:10.1097/01.gim.0000200948.58427.e2
36. Deignan JL, Astbury C, Cutting GR, et al. CFTR variant testing: a technical standard of the American College of Medical Genetics and Genomics (ACMG). Genetics in Medicine. 2020/08/01 2020;22(8):1288-1295. doi:10.1038/s41436-020-0822-5
37. Deignan JL, Gregg AR, Grody WW, et al. Updated recommendations for CFTR carrier screening: A position statement of the American College of Medical Genetics and Genomics (ACMG). Genetics in Medicine. 2023/08/01/ 2023;25(8):100867. doi:10.1016/j.gim.2023.100867
38. ACMG. Technical Standards and Guidelines for CFTR Mutation Testing. Standards and Guidelines for Clinical Genetics Laboratories. Mar 2011;
39. Gregg AR, Aarabi M, Klugman S, et al. Screening for autosomal recessive and X-linked conditions during pregnancy and preconception: a practice resource of the American College of Medical Genetics and Genomics (ACMG). Genetics in Medicine. 2021/10/01 2021;23(10):1793-1806. doi:10.1038/s41436-021-01203-z
40. Farrell PM, White TB, Ren CL, et al. Diagnosis of Cystic Fibrosis: Consensus Guidelines from the Cystic Fibrosis Foundation. The Journal of pediatrics. Feb 2017;181s:S4-S15.e1. doi:10.1016/j.jpeds.2016.09.064
41. Ren CL, Morgan RL, Oermann C, et al. Cystic Fibrosis Foundation Pulmonary Guidelines. Use of Cystic Fibrosis Transmembrane Conductance Regulator Modulator Therapy in Patients with Cystic Fibrosis. Annals of the American Thoracic Society. Mar 2018;15(3):271-280. doi:10.1513/AnnalsATS.201707-539OT
42. Green DM, Lahiri T, Raraigh KS, et al. Cystic Fibrosis Foundation Evidence-Based Guideline for the Management of CRMS/CFSPID. Pediatrics. 2024;153(5)doi:10.1542/peds.2023-064657
43. ACOG. Committee Opinion No. 691: Carrier Screening for Genetic Conditions. Obstetrics and gynecology. Mar 2017;129(3):e41-e55. doi:10.1097/aog.0000000000001952
44. Langfelder-Schwind E, Karczeski B, Strecker MN, et al. Molecular testing for cystic fibrosis carrier status practice guidelines: recommendations of the National Society of Genetic Counselors. Journal of genetic counseling. Feb 2014;23(1):5-15. doi:10.1007/s10897-013-9636-9
45. S+02myth AR, Bell SC, Bojcin S, et al. European Cystic Fibrosis Society Standards of Care: Best Practice guidelines. Journal of Cystic Fibrosis. 2014;13:S23-S42. doi:10.1016/j.jcf.2014.03.010
46. Castellani C, Duff AJA, Bell SC, et al. ECFS best practice guidelines: the 2018 revision. Journal of cystic fibrosis : official journal of the European Cystic Fibrosis Society. Mar 2018;17(2):153-178. doi:10.1016/j.jcf.2018.02.006
47. NICE. Cystic fibrosis: diagnosis and management. https://www.nice.org.uk/guidance/ng78/chapter/Recommendations#diagnosis-of-cystic-fibrosis
48. PCRM. Nutrition Guide for Clinicians: Cystic Fibrosis. Physicians Committee for Responsible Medicine. Updated February 16, 2023. https://nutritionguide.pcrm.org/nutritionguide/view/Nutrition_Guide_for_Clinicians/1342064/all/Cystic_Fibrosis
49. FDA. 510(k) Substantial Equivalence Determination Decision Summary. http://www.accessdata.fda.gov/cdrh_docs/reviews/K051435.pdf
50. FDA. Decision Summary Inplex CF Molecular Test 510(k) Premarket Notification. https://www.accessdata.fda.gov/cdrh_docs/reviews/K063787.pdf
51. FDA. Decision Summary eSensor® CF Genotyping Test 510(k) Premarket Notification. https://www.accessdata.fda.gov/cdrh_docs/reviews/K090901.pdf
52. FDA. Decision Summary xTAG® Cystic Fibrosis 60 Kit v2 510(k) Premarket Notification. https://www.accessdata.fda.gov/cdrh_docs/reviews/K083845.pdf
53. FDA. XTAG Cystic Fibrosis 60 Kit V2, XTAG Data Analysis Software (TDAS) CFTR. https://www.accessdata.fda.gov/cdrh_docs/pdf16/K163336.pdf
54. FDA. Decision Summary Illumina MiSeqDx™ Cystic Fibrosis 139-Variant Assay 510(k) Premarket Notification. https://www.accessdata.fda.gov/cdrh_docs/reviews/K124006.pdf
55. FDA. XTAG Cystic Fibrosis 39 Kit V2. https://www.accessdata.fda.gov/cdrh_docs/pdf16/K163347.pdf
Coding Section
| Code | Number | Description |
| CPT | 81220 | CFTR (cystic fibrosis transmembrane conductance regulator) (e.g., cystic fibrosis) gene analysis; common variants (e.g., ACMG/ACOG guidelines) |
| 81221 | CFTR (cystic fibrosis transmembrane conductance regulator) (e.g., cystic fibrosis) gene analysis; known familial variants | |
| 81222 | CFTR (cystic fibrosis transmembrane conductance regulator) (e.g., cystic fibrosis) gene analysis; duplication/deletion variants | |
| 81223 | CFTR (cystic fibrosis transmembrane conductance regulator) (e.g., cystic fibrosis) gene analysis; full gene sequence | |
| 81224 | CFTR (cystic fibrosis transmembrane conductance regulator) (e.g., cystic fibrosis) gene analysis; intron 8 poly-T analysis (e.g., male infertility) | |
| 81412 | Ashkenazi Jewish associated disorders (e.g., Bloom syndrome, Canavan disease, cystic fibrosis, familial dysautonomia, Fanconi anemia group C, Gaucher disease, Tay-Sachs disease), genomic sequence analysis panel, must include sequencing of at least 9 genes, including ASPA, BLM, CFTR, FANCC, GBA, HEXA, IKBKAP, MCOLN1, and SMPD1 | |
| 81479 | Unlisted molecular pathology procedure |
Procedure and diagnosis codes on Medical Policy documents are included only as a general reference tool for each policy. They may not be all-inclusive.
This medical policy was developed through consideration of peer-reviewed medical literature generally recognized by the relevant medical community, U.S. FDA approval status, nationally accepted standards of medical practice and accepted standards of medical practice in this community and other nonaffiliated technology evaluation centers, reference to federal regulations, other plan medical policies, and accredited national guidelines.
"Current Procedural Terminology © American Medical Association. All Rights Reserved"
History From 2017 Forward
| 07/28/2025 | Annual review, updating coverage criteria for clarity and consistency. New CC 6 for family members of individuals with CFTR-related metabolic disorder/cystic fibrosis screen positive, inconclusive diagnosis (CRMS/CFSID), comprehensive analysis of CFTR gene analysis. Also updating description, table of terminology, rationale, and references. |
| 08/12/2024 | Annual review, updating note 1 to include updated ACMG mutation list. Also updating rationale and references. |
| 08/10/2022 | Annual review, policy reformatted for clarity, adding coverage criteria #7. Also updating descrition, rationale and references. |
| 07/29/2021 |
Annual review, updating policy for specificity of #3 adn #4. No change to policy intent. |
| 07/22/2020 |
Annual review, no change to policy intent. Updating background, guidelines and references. |
| 07/12/2019 |
Annual review, no change to policy intent. |
| 06/19/2019 |
Interim review. Genetic counseling is recommended is replacing Genetic counseling is Medically necessary. No other changes made. |
| 07/18/2018 |
Annual review, no change to policy intent. |
| 10/09/2017 |
Updated Gene Testing for Cystic Fibrosis table. No other changes |
| 07/19/2017 |
New Policy |