Prenatal Testing for Fetal Aneuploidy - CAM 254HB
Description:
Aneuploidy is defined as an abnormal number of chromosomes present in the cell. Fetal aneuploidy is a condition where the fetus has one or more extra or missing chromosomes leading to either a nonviable pregnancy, offspring that may not survive after birth, or surviving newborn with congenital birth defects and functional abnormalities. The most common fetal aneuploidies associated with an additional chromosome are Down syndrome (trisomy 21), Edwards syndrome (trisomy 18), and Patau syndrome (trisomy 13). Prenatal screening for fetal aneuploidy is an assessment of the pregnant individual’s risk of carrying a fetus with fetal aneuploidy using markers found in maternal serum (ACOG, 2016). Noninvasive prenatal screening is a method for screening for chromosomal abnormalities using a maternal blood sample where cell-free fetal DNA (cffDNA) is extracted and screened for aneuploidies (McKanna et al., 2018).
For guidance on the use of chromosomal microarray in the confirmation of fetal aneuploidy, please see CAM 313 Chromosomal Microarray and Low-pass Whole Genome Sequencing.
Regulatory Status
In April 2021, the FDA released a statement titled, “Genetic Non-Invasive Prenatal Screening Tests May Have False Results: FDA Safety Communication,” that detailed safety concerns regarding the use of NIPS tests outside of standard screening purposes. Specifically, they reinforced that NIPS tests are laboratory developed tests not regulated by the FDA and that the use of NIPS should be confined to screenings, rather than diagnoses. “The accuracy and performance of NIPS tests have not been evaluated by the FDA and these tests can give false results, such as reporting a genetic abnormality when the fetus does not actually have one. NIPS tests are screening tests, which means the NIPS test may only tell you the risk of the fetus having certain genetic abnormalities. They are not diagnostic tests, which are generally used to more definitively confirm or rule out a suspected genetic abnormality.” Additionally, “The FDA recommends that patients discuss the benefits and risks of NIPS tests with a genetic counselor or other health care provider before deciding to get these tests. Patients should also discuss the results of NIPS tests with a genetic counselor or other health care provider before making any decisions about their pregnancy. Health care providers should be aware of the risks and limitations of using these screening tests and should not use the results from these tests alone to diagnose chromosomal (genetic) abnormalities or disorders” (FDA, 2022).
Many labs have developed specific tests that they must validate and perform in house. These laboratory-developed tests (LDTs) are regulated by the Centers for Medicare & Medicaid Services (CMS) as high-complexity tests under the Clinical Laboratory Improvement Amendments of 1988 (CLIA ’88). LDTs are not approved or cleared by the U.S. Food and Drug Administration; however, FDA clearance or approval is not currently required for clinical use.
Policy:
Application of coverage criteria is dependent upon an individual’s benefit coverage at the time of the request.
- For pregnant individuals who desire information on the risk of having a child with fetal aneuploidy, the following screening tests to detect fetal aneuploidy of chromosomes 13, 18, and 21 is considered MEDICALLY NECESSARY:
- First-trimester (defined as 11 – 14 weeks) screening incorporating maternal serum markers (hCG, PAPP-A with NT).
- Second-trimester (15 – 22 weeks) screening incorporating triple maternal serum markers (hCG, AFP, uE3 with NT) and quad maternal serum markers (hCG, AFP, uE3, DIA with NT).
- First (11 – 14 weeks) and second (15 – 22 weeks) trimester integrated screening incorporating maternal serum markers (PAPP-A with NT) and quad maternal serum markers (hCG, AFP, uE3, DIA with NT).
- First (11 – 14 weeks) and second (15 – 22 weeks) trimester sequential screening incorporating maternal serum markers (PAPP-A, hCG with NT) and quad maternal serum markers (hCG, AFP, uE3, DIA with NT).
- First (11 – 14 weeks) and second (15 – 22 weeks) trimester contingent screening incorporating maternal serum markers (PAPP-A, hCG with NT); if positive, quad maternal serum markers (hCG, AFP, uE3, DIA with NT).
- For pregnant individuals who desire information on the risk of having a child with fetal aneuploidy, non-invasive prenatal screening (NIPS) to detect fetal aneuploidy of chromosomes 13, 18, 21, X, and Y is considered MEDICALLY NECESSARY when all of the following criteria (a – d) are met:
- Underwent pretest counseling
- A cell-free fetal DNA test has not been performed yet in this pregnancy
- Current pregnancy is not a multiple gestation
- Current pregnancy is greater than or equal to 10 weeks and less than 23 weeks at the time the blood will be drawn
- For pregnant individuals who desire information on the risk of having a child with fetal aneuploidy or to pursue additional confirmatory testing of equivocal or positive results from the above testing, karyotyping to confirm fetal aneuploidy is considered MEDICALLY NECESSARY.
- To detect fetal aneuploidy, the use of the “penta” screen (hCG, AFP, uE3, DIA with NT, and hyperglycosylated hCG) is considered
NOT MEDICALLY NECESSARY.
- Screening for the detection of fetal aneuploidies is considered NOT MEDICALLY NECESSARY in any of the following situations:
- Parallel or simultaneous testing with multiple screening methodologies for fetal aneuploidy.
-
- For the screening of pregnant individuals with higher order multiple gestation pregnancies.
-
- Repeat screening for pregnant individuals with negative screening results.
-
- For the detection of other chromosomal abnormalities (e.g., microdeletion syndromes, unbalanced translocations, deletions, duplications) not addressed above.
- For the determination of fetal sex.
The following does not meet coverage criteria due to a lack of available published scientific literature confirming that the test(s) is/are required and beneficial for the diagnosis and treatment of an individual’s illness.
- For the diagnosis of fetal aneuploidy, the use of single cell genotyping in trophoblasts isolated from maternal serum (e.g., Luna Prenatal Test) is considered NOT MEDICALLY NECESSARY.
Table of Terminology
| Term |
Definition |
| ACOG |
The American College of Obstetrics and Gynecologists |
| ACMG |
The American College of Medical Genetics and Genomics |
| AFP |
Alpha-fetoprotein |
| ASRM |
American Society for Reproductive Medicine |
| BMI |
Body mass index |
| cfDNA |
Cell-free deoxyribose nucleic acid |
| CffDNA |
Cell-free fetal deoxyribose nucleic acid |
| CGH |
Comparative genomic hybridization |
| CLIA ’88 |
The Clinical Laboratory Improvement Amendments of 1988 |
| CMA |
Chromosomal microarray |
| CMS |
Centers for Medicare & Medicaid Services |
| CPM |
Confined placental mosaicism |
| CVS |
Chorionic villa sampling |
| DIA |
Dimeric inhibin A |
| DNA |
Deoxyribose nucleic acid |
| dPCR-NIPT |
Droplet digital polymerase chain reaction-non-invasive prenatal testing |
| E3 |
Estriol |
| FASTER |
First and second trimester evaluation of risk |
| FDA |
Food and Drug Administration |
| FPR |
False-positive rate |
| GCPG |
Practice Committee and Genetic Counseling Professional Group |
| β-hCG |
β-human chorionic gonadotropin |
| hCG |
Human chorionic gonadotropin |
| h-hCG |
Hyperglycosylated human chorionic gonadotropin |
| Ifs |
Incidental findings |
| ISPD |
The International Society for Prenatal Diagnosis |
| ITA |
Invasive trophoblast antigen |
| LDT |
Laboratory-developed test |
| MPS |
Massively parallel sequencing |
| MSS |
Maternal serum screening |
| NEXT |
Noninvasive examination of trisomy |
| NGS |
Next generation sequencing |
| NIPD |
Noninvasive prenatal diagnosis |
| NIPS |
Noninvasive prenatal screening |
| NIPT |
Noninvasive prenatal testing |
| NSGC |
The National Society of Genetic Counselors |
| NT |
Nuchal translucency |
| PAPP-A |
Pregnancy-associated plasma protein-A |
| PPV |
Positive predictive value |
| RATs |
Rare autosomal trisomies |
| SCAs |
Sex chromosome aneuploidies |
| SMFM |
Society for Maternal-Fetal Medicine |
| SNPs |
Single nucleotide polymorphisms |
| SURUSS |
Serum, Urine and Ultrasound Screening Study |
| uE3 |
Unconjugated estriol |
Rationale
Pregnant individuals are routinely offered blood-based screening or invasive diagnostic testing for identification of the most common fetal aneuploidies: trisomy 13 (Patau syndrome), trisomy 18 (Edwards syndrome) and trisomy 21 (Down syndrome). Approximately 90% of chromosomal abnormalities are due to an incorrect number of chromosomes, especially in these three triploid conditions as well as monosomy X (Turner syndrome). Approximately 15% – 20% of clinically recognized pregnancies result in first trimester spontaneous abortions, with 50% of these spontaneous abortions due to chromosomal abnormalities (Witters et al., 2011).
Historically, noninvasive blood-based aneuploidy screening has taken the form of first- and/or second-trimester analysis of biomarkers in maternal circulation, sometimes along with ultrasound measurement of fetal nuchal translucency (NT). Although both the sensitivity (detection rate) and specificity (true positive rate) of maternal serum screening tests for aneuploidy have improved significantly over time, the false positive rate (two to five percent) remains higher than desirable. The detection rate of Down syndrome in the first trimester using a combination of NT and biochemical markers is typically 79% – 90% (Dey et al., 2013). Positive maternal serum screen results are usually followed by an invasive diagnostic test, such as karyotyping of a chorionic villus sample (in first trimester) or karyotyping of an amniotic fluid sample (second trimester).
Additionally, detection rates of maternal serum screens are typically below 99%, resulting in the inability of a normal result to confer complete confidence that the fetus is unaffected with aneuploidy (Dey et al., 2013). Thus, many pregnant individuals who are in a high-risk category due to age or other factors may opt for the more definitive, diagnostic, invasive testing, which has its own risks and relatively high costs. The availability of noninvasive testing may improve both the sensitivity and specificity of aneuploidy detection while resulting in fewer invasive procedures, less risk, and less overall cost.
Screening Tests
Chromosomal anomalies are a leading cause of perinatal mortality and developmental abnormality. The goal of prenatal testing is to screen for chromosomal anomalies and to provide genetic counseling for parents. The American College of Obstetrics and Gynecology (ACOG) recommends that prenatal testing is offered to all pregnant individuals (ACOG, 2016, 2020). Invasive testing, including chorionic villi sampling or amniocentesis, should be limited to high-risk patients owing to the potential risks for procedure-related pregnancy loss.
Genome-wide sequencing tests for fetal diagnostics have also been employed and are expected to increase in popularity as the cost decreases and as new tools are developed. These tests include DNA sequencing methods, such as whole exome-sequencing and targeted clinical panels, which can further evaluate fetal structural anomalies first detected in an ultrasound (ISPD, 2018). This diagnostic sequencing method has been used for various fetal diagnostic measures including standard genetic testing and chromosomal microarray analysis. While cfDNA could theoretically be analyzed to screen for other genetic disorders beyond common aneuploidies, no professional guideline currently recommend expanded screening for additional genetic disorders (Glenn E Palomaki, 2024). Screening for these additional genetic conditions (other aneuploidies, microdeletions/microduplications, and single gene disorders) while increasingly commercially available, has not been recommended for routine use by leading medical societies and is considered “investigational” (Glenn E Palomaki, 2024).
Chromosomal microarray (CMA) testing refers to the use of comparative genomic hybridization (CGH) arrays to compare the DNA of a patient with a normal control (Aradhya et al., 2007). CMA is significantly more sensitive (10 to 100 kb) than traditional karyotyping (5 to 10 Mb) and has a turnaround time of five days quicker than karyotyping (Robson et al., 2017), while providing an alternative to karyotyping when dividing cells are not available for analysis. This technique may be used for several different purposes, such as identifying a cause of pregnancy loss or identifying other aneuploid conditions, such as Down syndrome (Reddy et al., 2012). This method of diagnostic prenatal sequencing is currently investigational because of limited data and is utilized most prominently in research settings or clinically on a case-by-case basis (ISPD, 2018).
Biochemical Markers in Maternal Serum
Many studies revealed that maternal age, fetal NT, maternal serum free β-human chorionic gonadotropin (hCG) and pregnancy-associated plasma protein-A (PAPP-A) have been associated with aneuploidy. The "Quad screen", comprising alpha-fetoprotein (AFP), hCG, unconjugated estriol (E3), and inhibin A, is the most efficient multiple-marker screening test in the second trimester. In addition, there are more options such as integrated, sequential testing, and cell-free DNA screening. Many studies are ongoing to reveal the most sensitive, specific and effective screening tools for use during the first trimester (Park et al., 2016).
To improve the accuracy of serum markers, ultrasound markers are used. NT refers to the fluid filled space measured on the dorsal aspect of the fetal neck. An enlarged NT (>3.0 mm/99th percentile of the crown-rump length) is independently associated with fetal aneuploidy and structural malformations (ACOG, 2016, 2020).
Screening studies of pregnant individuals reported an association between increased NT in the first trimester of pregnancy (10-13 weeks of gestation) and chromosomal defects, most commonly Down syndrome (trisomy 21) but also trisomy 18 and 13. NT could be done alone as a first-trimester screen or in combination with maternal serum markers, free beta subunit of human chorionic gonadotropin (β-hCG) and pregnancy-associated plasma protein-A (PAPP-A). All three trisomies (chromosomes 13, 18, and 21) “are associated with increased maternal age, increased fetal NT and decreased PAPP-A, but in trisomy 21 serum free β-hCG is increased whereas in trisomies 18 and 13 free β-hCG is decreased” (Shiefa et al., 2013). Low β-hCG in the first trimester has also been associated with an increased risk of significant copy number variants on chromosomal microarray analyses (Bornstein et al., 2018).
Analytical Validity of Biochemical Markers
Screening for chromosomal abnormalities using biochemical markers include the first trimester combined test, triple test, quadruple test, sequential test, and integrated test. Except for the first trimester combined test, all others can provide screening results in the second trimester. In the first trimester combined test, the risk is calculated based on the ultrasonographic findings of NT and maternal serum levels of free β-hCG and PAPP-A. First-trimester screening not only allows early reassurance or early diagnosis of aneuploidy, but it also provides an option of earlier and safer termination of pregnancy in affected cases. Consequently, the first trimester combined test has become one of the most popular and useful screening strategies. Lee et al. (2013) conducted a 13-year study of 25,104 pregnant individuals using the first trimester Down syndrome screening. “The detection rates for trisomy 21, trisomy 18, Turner syndrome, and other chromosome anomalies were 87.5% (21/24), 69.2% (9/13), 81.8% (9/11), and 60% (18/30), respectively, with a false positive rate (FPR) of 5.4% (1353/25,026). Further evaluation of the detection rates for trisomy 21, by gestational age at 11, 12, and 13 weeks, were 92.3%, 87.5%, and 66.7%, respectively” (Lee et al., 2013).
For second-trimester screening for Down syndrome, the sensitivity and specificity of the triple test — co-testing AFP, unconjugated E3, and free β-hCG — are higher than screening with AFP alone. However, when the false-positive rate is fixed at five percent, to compare the screening performance between the screening tools, the detection rate was found to be 66.8% to 77% with the triple test and 75.9% to 92% with the first trimester combined test. The sensitivity of the triple test was lower than the combined test (Baer et al., 2015).
The quadruple test, which uses the fourth marker, inhibin A, in addition to the other three markers, has seven percent higher sensitivity when applying a fixed five percent false-positive rate. A study conducted by Wald et al. (2003) revealed that when inhibin A was added to the traditional triple marker test, a detection rate of 83% was achieved, which was six percent higher than the 77% detection rate found with the triple test. This result was similar to that produced with the first trimester combined test (Park et al., 2016).
Many studies, including the Serum, Urine and Ultrasound Screening Study (SURUSS) (Wald et al., 2003) and the First and Second-Trimester Evaluation of Risk (FASTER) study (Malone et al., 2005), have offered evidence suggesting that first-trimester screening for Down syndrome with measurement of fetal NT and maternal serum markers is at least as accurate as alternative tests and may allow for earlier confirmation or exclusion of Down syndrome. These studies evaluated several tests in parallel, including first trimester testing with NT and maternal markers, the triple test, second-semester quadruple test and a combined first- and second-trimester test (both with and without NT), stepwise sequential testing (results given after first-trimester testing, move on to second-trimester testing), and integrated screening (results given only after first and second-trimester testing). In a direct comparison of the first-trimester test to the triple test, the SURUSS study has shown that setting the false-positive rate at 5% resulted in an 83% detection rate, which was superior to what was historically expected of the triple test (Wald et al., 2003). SURUSS results were based on data from 47,053 pregnancies (101 with Down syndrome). The FASTER trial was conducted in the United States and was sponsored by the National Institutes of Health. The study enrolled 38,167 pregnant individuals and provided further evidence that first-trimester combined screening was effective, but it did not provide NT measurement alone; results showed that integrated first- and second-trimester screening provided higher detection rates. The SURUSS and FASTER studies also found that overall, first-trimester screening with NT alone is inferior to either first- or second-trimester combined screening. Additional testing may not be necessary in those few cases when NT is at least 4.0 mm due to the high likelihood of Down syndrome in these cases (Malone et al., 2005; Wald et al., 2009; Wald et al., 2003).
Studies have found a high rate of successful imaging of the fetal nasal bone and an association between absent nasal bone and the presence of Down syndrome in high-risk populations. However, there is insufficient evidence on the performance of fetal nasal bone assessment in average-risk populations. Of concern is the low performance of fetal nasal bone assessment in a subsample of the FASTER study conducted in a general population sample. Two studies conducted outside of the United States have found that, when added to a first-trimester screening program evaluating maternal serum markers and NT, fetal nasal bone assessment can result in a modest decrease in the false-positive rate. Several experts in the field are proposing that fetal nasal bone assessment be used as a second stage of screening to screen pregnant individuals found to be of borderline risk using maternal serum markers and NT. Considering the uncertainty of test performance in average-risk populations and the lack of standardization in the approach to incorporating this test into a first-trimester screening program, detection of fetal nasal bone is considered investigational (Wald et al., 2009).
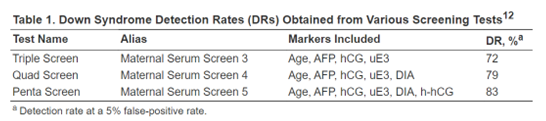
Palomaki et al. (2004) demonstrate that hyperglycosylated hCG (h-hCG), also known as invasive trophoblast antigen (ITA), may be a promising screening marker for Down syndrome detection in the second trimester. In the study, serum samples of 45 Down syndrome cases and 238 unaffected control pregnancies between 14 to 20 weeks of gestation were collected and measured for h-hCG, along with other screening markers (Palomaki et al., 2004). As seen in the figure below, h-hCG, in combination with four other screening markers, increased the detection rate to 83% at a 5 percent false-positive rate from the 72% detection rate by the tripe test (Palomaki et al., 2004; QuestDiagnostics, 2022). In addition, “The median [h-hCG] in Down syndrome pregnancies was > 3.00 multiples of the median, higher than that found for human chorionic gonadotropin (hCG).” The author recommends that “the highest screening performance for Down syndrome can be obtained by integrating first- and second-trimester serum and ultrasound markers into a single interpretation in the second trimester. This integrated test approach can detect 90% of Down syndrome pregnancies at a 3% false-positive rate” (Palomaki et al., 2004).
Cell-Free Fetal DNA from Maternal Serum
In 1997, researchers reported the identification of cell-free fetal DNA (cffDNA) in the circulation of pregnant individuals (Lo et al., 1997). The fetal fraction of cffdna increases throughout gestation. cffDNA is usually detectable within six to seven weeks of gestation, with the earliest detection reported at 4.5 weeks. Therefore, it allows for noninvasive procedures to be performed much earlier in the pregnancy and eschews the need for standard biochemical and invasive screens. Moreover, given that cffDNA is cleared from maternal circulation within hours following birth and removal of the placenta, cffDNA is specific to the ongoing pregnancy at the time of sampling (Shaw et al., 2020). At 10 weeks of gestation, the fetal fraction comprises at least three to four percent whereas it can account for up to 50% of the total cell-free DNA at term (Palomaki et al., 2024). Since then, laboratories have validated several different techniques for the use of cell-free DNA as a screening test for fetal aneuploidy, and these methods have been termed noninvasive prenatal screening (NIPS) or noninvasive prenatal testing (NIPT).
Noninvasive prenatal screening is a testing method which utilizes cell-free DNA from the plasma of pregnant individuals to screen for fetal aneuploidy. It is important to note that cell-free DNA screening does not assess risk of fetal anomalies, including neural tube defects or ventral wall defects (ACOG, 2015). NIPS methods only provide an estimate of whether the risk of aneuploidy is increased or decreased; NIPS does not provide a definitive diagnosis of aneuploidy. As with other aneuploidy screening tests, it is recommended that positive results of NIPS be followed by diagnostic testing such as traditional karyotyping of fetal cells obtained via chorionic villus sampling or amniocentesis (Gregg et al., 2016).
One cell-free fetal DNA detection method for NIPS, known as massively parallel sequencing (MPS), is a technique in which millions of pieces of maternal and fetal chromosomal material are sequenced and quantified. The MPS method is able to detect many types of aneuploidies, including those which are less commonly seen (Devers et al., 2013). MPS can detect common aneuploidies with both high sensitivity and high specificity for trisomies 13, 18, and 21. Bianchi et al. (2012) found the detection rate sensitivity for trisomy 21 to be 100%, the detection rate sensitivity for trisomy 18 to be 97.2%, and the detection rate sensitivity for trisomy 13 to be 78.6%; specificity was 100% for all three of the aforementioned trisomies.
Detection of aneuploidy using circulating cell-free fetal DNA can also be performed using selective analysis of specific loci only from the chromosomes of interest, as opposed to sequencing of all chromosomes performed in MPS. This directed analysis of cell-free fetal DNA has also been shown to have high sensitivity and high specificity for the common trisomies. Lee et al. (2019) utilized plasma from 1,055 pregnant individuals and found that NIPT with cell-free fetal DNA “showed 100% sensitivity and 99.9% specificity for trisomy 21, and 92.9% sensitivity and 100% specificity for trisomy 18, and 100% sensitivity and 99.9% specificity for trisomy 13” (Lee et al., 2019).
The third approach to detect aneuploidy from cffdna is based on the amplification of single nucleotide polymorphisms (SNPs) on the chromosome of interest. In a study by Eiben et al. (2015), 2,942 patients underwent SNP-based noninvasive prenatal screening (NIPS) in which the source for cffDNA was derived from placental cells. Sixty-five patients (2.2%) had positive noninvasive prenatal screening results for aneuploidy and further invasive testing confirmed aneuploidy in fifty-nine of those patients (90.8%). The remaining six patients were false positives due to a discrepancy between the genetic status of the fetus and placenta, a condition known as confined placental mosaicism (CPM). The fetal fraction was abnormally low (less than eight percent) and indicative of fetal-placental discrepancies. Although a reliable screening method, the author suggests that SNP-based NIPS “cannot be used as a standalone test without ultrasound examination or invasive confirmation” (Eiben et al., 2015).
Despite the apparent advantages of NIPS over standard maternal serum screening in screening for common aneuploidies, there are limitations. “Reported Ifs [incidental findings] range from fetal or maternal deletions and duplications or mosaic sex chromosome aneuploidy in the mother or fetus, presenting as aneuploidy risk on NIPS, to mosaicism and uniparental disomy to abnormal results because of the presence of cell-free DNA originating from an undiagnosed maternal tumor” (Westerfield et al., 2014). When ultrasound evaluation reveals fetal anomalies that may be consistent with one of those scenarios, invasive diagnostic testing with karyotyping or microarray may be more appropriate. NIPS also cannot distinguish the cause of aneuploidy, nor can it differentiate among the presence of an extra chromosome, a Robertsonian translocation, or high-level mosaicism. The determination of the type of aneuploidy is important for accurate counseling and future risk assessment (Neufeld-Kaiser et al., 2015; Strom et al., 2017; Westerfield et al., 2014). Also, some samples contain insufficient amounts of cell-free DNA, which is unknown until the test procedure has commenced. Early gestational age (<10 weeks) and high body mass index have been shown to be associated with reduced amounts of circulating cell-free fetal DNA. Additionally, NIPS for aneuploidy does not detect the presence of neural tube defects, which is included in traditional second trimester maternal serum screening. It has been suggested that the testing of maternal serum AFP in the second trimester should be offered to pregnant individuals who underwent first-trimester aneuploidy screenings (Palomaki et al., 2024).
And so, while promising on the screening front, research has yet to support NIPT’s diagnostic prowess. NIPT platforms typically screen for common trisomies with or without sex chromosome anomalies, and therefore overlook most other chromosomal rearrangements (Al Toukhi et al., 2019; Shaw et al., 2020). Furthermore, the power of NIPT is limited by discordant — e.g., false-positive and false-negative — results, due to issues including vanishing twin syndrome, where a spontaneous early miscarriage may still release cffDNA and interfere with early NIPT results. Abnormal maternal cells mixing with normal fetal cells, producing mosaicism as aforementioned, has been reportedly repeatedly and therefore is an incidental cause of discordant results, suggesting that pregnant individuals with known malignancies should be dissuaded from NIPT (Bianchi et al., 2015; Shaw et al., 2020).
Extension of NIPT to sex chromosome aneuploidies and rare autosomal trisomies has also been explored, though its utility remains controversial. The increased variability in its use here is due in part to the sensitivity of NIPT to detect sex chromosome aneuploidies — e.g., Turner syndrome (45, X) and Klinefelter syndrome (45, XXY) — being lower than that of common trisomies. Moreover, as NIPT screens were originally limited in scope to identify trisomies 13, 18, and 21, the utility of NGS-based NIPT to also detect rare autosomal trisomies (RATs) has yet to be informed by the clinical community, and offers inspiration for future directions (Shaw et al., 2020).
Analytical Validity of Cell-free Fetal DNA Testing
A study by Palomaki et al. (2012) of 4664 pregnancies at high-risk for Down syndrome using the MPS method had a detection rate of 98.6% with a false-positive rate of only 0.20% (3/1471) (Palomaki et al., 2011). Results also identified MPS as a successful detection method for both trisomy 18 and trisomy 13. The authors state, “Among the 99.1% of samples interpreted (1,971/1,988), observed trisomy 18 and 13 detection rates were 100% (59/59) and 91.7% (11/12) at false-positive rates of 0.28% and 0.97%, respectively. … Among high-risk pregnancies, sequencing circulating cell-free DNA detects nearly all cases of Down syndrome, trisomy 18, and trisomy 13, at a low false-positive rate. This can potentially reduce invasive diagnostic procedures and related fetal losses by 95%. Evidence supports clinical testing for these aneuploidies” (Palomaki et al., 2012).
Norton et al. (2015) reported near-perfect accuracy of detection for trisomy 21 (Down syndrome) with the use of cell-free DNA (cfDNA) (sensitivity, 100% [38 of 38 cases of trisomy 21]; false-positive rate, 0.06% [9 false positives among 15,841 pregnant individuals]) in the Noninvasive Examination of Trisomy (NEXT) study. Norton and colleagues found that cfDNA testing for trisomy 21, as compared with standard screening, had a better global performance during the first trimester of pregnancy. However, they did not provide information about the 14 fetal chromosomal abnormalities in the 15,841 screened pregnancies, other than for trisomies 13, 18, and 21 (Norton et al., 2015).
In 2017, the Dutch Ministry of Health introduced a nationwide implementation study on NIPT as a first-tier strategy offered to all pregnant individuals in the TRIDENT-2 study. TRIDENT-2 was specific in its scope, as it excluded pregnancies with a vanishing or dischorionic twin, fetal ultrasound including a nuchal translucency greater than or equal to 3.5mm, or gestational age less than 11 weeks. Moreover, pregnant individuals with a history of being high-risk for the common trisomies and who have had an organ transplant were excluded as well, as were pregnant individuals with malignant neoplasia. Of all pregnancies that year, 73,239 (42%) opted for NIPT, it was found that though the number of common trisomies 13, 18, and 21 detected by NIPT was comparable to those of earlier studies, PPVs were higher than expected (53% PPV, 98%, 96%, respectively) with high sensitivities (100%, 91%, 98%, respectively), as confirmed by invasive prenatal testing or by postnatal bloodwork (van der Meij et al., 2019). However, the researchers do acknowledge potential limitations, namely not having presented data on sex chromosome aneuploidies and using different sequencing methods (e.g., NextSeq vs. HiSeq) and fetal fraction benchmarks for rejection across their three testing centers. However, despite issues to external validity, the authors conclude that “this study has confirmed that genome-wide NIPT is a reliable and robust screening test for the detection of fetal trisomies 21, 18, and 13” as they urge further research on screening for fetal pathology and adverse pregnancy outcomes (van der Meij et al., 2019).
As a secondary finding of the TRIDENT-2 study, NIPT may play a future role in raising suspicion of rare maternal malignancies. The TRIDENT-2 study enrolled 231,896 pregnant patients with NIPT results and showed incidental findings of malignancy-suspicious-NIPT in a small proportion of patients (Heesterbeek et al., 2022). The most common malignancies in individuals capable of becoming pregnant who are also of reproductive age include breast, cervical, ovarian, and colorectal cancers; leukemia; Hodgkin and non-Hodgkin lymphoma; thyroid cancer; and melanoma. When tumor cfDNA is incidentally found in maternal blood, there are no current professional guidelines that address the clinical management of cfDNA results. While only (0.02%) of NIPT results in the Heesterbeek et al. (2022)study showed indications of a maternal malignancy, researchers pointed out this information has clinical significance and can prompt a diagnostic workup through magnetic resonance imaging and computed tomography to enable counseling and confirmative diagnosis in these rare instances (Heesterbeek et al., 2022).
Luo et al. (2021) aimed to explore the efficacy of using NIPT to predict sex chromosome aneuploidies (SCAs) in a 34,717-patient sample study in China. Of the clinical pregnancies examined, 229 (0.66%) were associated with sex chromosome aneuploidies, with 78 of the cases reporting positive for 45,X and 151 sex chromosome trisomies (47,XXX, 47,XXY, 47XYY). In total, 193 of the 229 NIPT positive results acquiesced to confirmatory invasive prenatal diagnosis via karyotyping analysis of amniotic fluid and fluorescent in situ hybridization, and it was found that only 67 (34.7%) were true positives. The authors reported similarly low PPVs, with 23.07% for 45,X and 36%, 50%, and 27.27% for 47,XXX, 47,XXY, 47XYY, respectively. Given this performance of the NIPT, the authors concluded that “Confirmatory testing of abnormal results is recommended prenatally or after birth,” insinuating the current impotency of NIPT (Luo et al., 2021).
A two-year longitudinal study which utilized 11,414 material blood samples for NIPT. Among the 11,213 cases with confirmatory results, “94 T21, 39 T18, 8 T13, 15 XO, 6 XXX, 3 XYY, 5 XXY and 11,042 euploid cases were detected.” Overall, the sensitivity of NIPT was 98.90% (T21), 100.00% (T18), 100.00% (T13), 90.91% (XO), 100.00% (XXX), 100.00% (XYY), and 100.00% (XXY), while the specificity was 99.96% (T21), 99.97% (T18), 99.99% (T13), 99.96% (XO), 99.98% (XXX), 100.00% (XYY), and 99.99% (XXY). The authors concluded that “NIPT showed excellent performance as screening test for the detection of fetal T21, T18, T13 and SCA [sex chromosome aneuploidies] in mixed-risk pregnancies in Iran” (Garshasbi et al., 2019).
A study out of the Illumina laboratory (formerly Verinata) compared NIPS to standard maternal serum screening in pregnant individuals at average-risk for fetal aneuploidy. Their report included data of 5,974 samples tested for trisomies 13, 18, and 21 as well as monosomy X. Aneuploidy was detected in 4.8% of samples with only 0.2% putative false-positives and 0.08% false-negatives; however, 2.8% of cases had indefinite results for a single chromosome (Futch et al., 2013). Illumina more recently reported a more extensive study consisting of 85,298 clinical cases. “Aneuploidy was detected or suspected in 2,142 (2.5%) samples. For aneuploidy detected cases with known clinical outcomes, the overall positive predictive value (PPV) was 83.5% (608/728); observed PPVs for trisomies 21, 18, and 13 ranged from 50.0 to 92.8%” (Taneja et al., 2016).
Liang et al. (2019) used cell-free plasma DNA to assess the clinical utility of using an expanded noninvasive prenatal screening (“NIPS-Plus”) to detect aneuploidy and genome-wide microdeletion/microduplication syndromes (MMS). Of the 94,085 individuals with singleton pregnancies enrolled in the study, 1,128 were suspected of having clinically significant fetal chromosomal abnormalities. Follow-up testing in the study reported the positive predictive values (PPVs) of 95%, 82%, 46%, 29%, and 47% for T21, T18, T13, rare trisomies, and sex chromosome aneuploidies, respectively. For known MMS (n=32), PPVs were 93% (DiGeorge), 68% (22q11.22 microduplication), 75% (Prader-Willi/Angelman), and 50% (Cri-du-Chat). Thus, the researchers conclude that “the data have potential significance in demonstrating the usefulness of cfDNA profiling” and that “NIPS-Plus can be used as a first-tier pregnancy screening method to improve detection rates of clinically significant fetal chromosome abnormalities” (Liang et al., 2019).
A retrospective study by Wu et al. (2020) compared positive noninvasive prenatal screening (NIPS) results for aneuploidy to standard diagnostic tests such as traditional karyotyping and chromosomal microarray analysis (CMA). The study enrolled 551 pregnant individuals who screened positive for trisomy 13, trisomy 18, trisomy 21, and other sex chromosomal aneuploidies. Samples were obtained from either amniotic fluid or fetal cord blood and subsequent karyotyping or CMA confirmed a total of 256 out of 551 cases (46.4%) to possess chromosomal abnormalities concordant or partially concordant with NIPT results. Placental biopsies were obtained to assess the etiology of NIPS false positives and confined placental mosaicism (CPM) was found in 60% of the biopsies. The authors also reported that pregnant individuals with advanced maternal age (>35 years) had the highest positive predictive value (PPV) for trisomy 21 (87.8%), trisomy 18 (59.3%), and trisomy 13 (37.5%), while the PPV was significantly lower for pregnant individuals with young maternal age (<34 years) for trisomy 21 (71.9%), trisomy 18 (0%), and trisomy 13 (16.7%). This suggests that NIPS performs better in predicting aneuploidies for pregnancies with advanced maternal age than for pregnancies with young maternal age. However, the author notes that the PPVs showed “no significant upward trend when compared based on specific age categories (an interval of 5 years), which suggested that NIPT-positive result deserves equal attention from both providers and patients regardless of maternal age” (Wu et al., 2020).

Muzzey et al. (2020) studied and compared the sensitivities of both traditional aneuploidy screening and customized whole genome sequencing NIPS for pregnant individuals with high body mass index (BMI). Generally, pregnant individuals with high BMI have reduced fetal fraction during cf-DNA based NIPS, causing up to 24.3% of tests to fail. The study included 58105 patients, and the authors analyzed fetal fraction, aneuploidy, and BMI. Overall, across all obesity classes, and assuming no fetal fraction related rest failures, “analytical sensitivity for the investigated NIPS exceeded that of traditional aneuploidy screening for trisomies 13, 18, and 21.” The authors conclude that customized NIPS with “high accuracy at low FF and a low test-failure rate” is preferable over traditional aneuploidy screening for individuals with high BMI (Muzzey et al., 2020).
Dar et al. (2022) investigated the performance of cell-free DNA screening against a genetic confirmation of results with the goal of analyzing test performance and test failure (no-call rates). A total of 20,194 pregnant individuals were enrolled with a median gestational pregnancy of 12.6 weeks. The results of the genetic test were confirmed for 17,851 cases (88.4%). Among these were 13,043 low-risk and 4808 high-risk cases for aneuploidy. A total of 133 trisomies were diagnosed. These were composed of 100 (trisomy 21), 18 (trisomy 18) and 15 (trisomy 13). The positive rate of cfDNA screens was lower in the low-risk group as compared to the high-risk cohort (0.27% versus 2.2% P < .0001). Sensitivity and specificity were comparable between the two risk groups. The PPV for the low- and high-risk groups was 85.7% versus 97.5%, respectively. There were also 602 individuals who had a “no-call” outcome post-draw and 287 (1.61%) after a second draw. The authors concluded that in pregnant individuals “at a low risk for aneuploidy, single-nucleotide polymorphism-based cell-free DNA has high sensitivity and specificity, positive predictive value of 85.7% for trisomy 21 and 74.3% for the three common trisomies.” They also noted that “patients who receive a no-call result are at an increased risk of aneuploidy and require additional investigation" (Dar et al., 2022).
Dai et al. (2022) studied the sensitivity and specificity of a multiplex droplet digital PCR NIPT (dPCR-NIPT) assay. The authors propose that this test has a higher sensitivity than traditional serum biochemical tests and has a lower cost than NGS-NIPTs. The clinical performance of the dPCR-NIPT assay was tested with 238 maternal plasma samples, 36 of which were trisomy positive. “The dPCR-NIPT assay demonstrated a detection sensitivity of 100% and a specificity of 95.12%.” The authors noted that this is the first study done on the dPCR-NIPT assay but concluded that it is “superior” to traditional serum biochemical tests and NGS-NIPTs (Dai et al., 2022).
Proprietary Testing
Several methods for detection of fetal aneuploidy by analysis of circulating cell-free fetal DNA are commercially available. All have been validated in pregnancies deemed to be at high-risk for aneuploidy. Evaluation of this technology for use in low- or average-risk pregnancies is ongoing.
Current commercially available laboratory-developed noninvasive prenatal tests for aneuploidy include: the MaterniT21™ Plus Test(Integrated Genetics/LabCorp) (LabCorp, 2023a), the verifi™ Prenatal Test and verifi™ Plus Prenatal Test (Illumina) (Illumina, 2023), the Harmony Prenatal Test™ (Roche) (Roche, 2023), the Panorama test (Natera, available from several reference laboratories) (Natera, 2023), QNatal® Advanced test (QuestDiagnostics, 2023), Prequel test (Myriad, 2023), CentoNIPT® test (Centogene, 2023), ClariTestTM Core (GenPath, 2023), IONA® test and Sage™ prenatal screen (YourgeneHealth, 2023), Invitae NIPS test (Invitae, 2023), and Clarigo test (AgilentTechnologies, 2022). Other examples include, but are not limited to, the VERACITY® test out of NIPD Genetics (NIPD, 2023), the Vanadis® NIPT system (PerkinElmer, 2023), the NIFTY® Test and NIFTY® Test Pro (BGI, 2023), and the informaSeq® Prenatal Test (Genetics, 2016).
Regarding serum screening options for common birth defects, Integrated Genetics, a LabCorp Specialty Testing Group, names the Afp4®, which screens for Down syndrome, trisomy 18, and open neural tube defects in the second trimester, and SerumIntegratedScreen®, which screens for the same, but combines results from both the first and second trimesters. Other tests mentioned include the FirstScreen®, IntegratedScreen®, and SequentialScreen®, the latter of which boasts that “Part 1 [between 10th and 14th weeks of pregnancy] leads to the detection of approximately 70% of Down syndrome cases and 80% of trisomy 18 cases, and Part two [between 15th and 22nd weeks of pregnancy] leads to detection of approximately 90% of Down syndrome cases, 90% of trisomy 18 cases and 80% of open neural tube defects” (LabCorp, 2023b).
The Luna Prenatal Test is a prenatal genetic test that uses a cell-based approach to extract fetal DNA from fetal trophoblast cells present in maternal blood (Luna, 2022a). A key advantage of this test, highlighted in a white paper, is its ability to accurately analyze a single fetal cell ensuring access to pure fetal DNA.
In an analytical validation overview of the Luna Prenatal Test, two groups of pregnant individuals (one group composed of 59 high-risk individuals who received CVS or amniocentesis) and a second group (158 low-risk individuals who had no known diagnostic testing) were recruited. A third set of test samples was analyzed for comparison composed of “seven Coriell cell lines harboring known chromosome abnormalities” (Luna, 2022b). Single cells (enriched from maternal whole blood) were processed individually to access pure fetal or cell line DNA. Subsequently, the DNA from each individual cell was “sequenced by low-pass massively parallel sequencing technology to generate a genome-wide copy number profile, which was evaluated for genome-wide copy number changes larger than 1.5 Mb”(Luna, 2022b). The first group of 59 samples had 11 samples excluded. Of the remaining 48 pregnancies (49 fetuses), results indicated an accuracy of 100% (92.75% – 100% confidence interval), specificity 100% (54.07% – 100% confidence interval), sensitivity of 100% (91.78% – 100% confidence interval), PPV of 100%, and NPV of 100% in detecting chromosomal abnormalities (complete agreement between CVS/amniocentesis and Luna); they also noted a plan to increase the sample size to 200 for a larger data set as this was a limited study.
For the second test group, a report was issued for 91.1% of cases, with 8.9% receiving a result of “no scorable cells.” In the second group validation study, they found “complete intra-case concordance for 404 cells from 118 cases” (Luna, 2022b). The third set used human cultured cells (lymphoblast or fibroblast) with known aneuploidy or deletion. From the results: “out of a total of 148 scorable cells, the known abnormality was detected and called by the NxC software in every cell except one. Thus, the analytic sensitivity and specificity were 100% with the expected finding being reported in every cell” (Luna, 2022b).
The test makers also note: “We do not claim that the Luna Prenatal Test is diagnostic for aneuploidy, because there are recommendations from professional organizations that a final diagnosis should not rest on testing of trophoblasts alone” (Luna, 2022b).
Bellair et al. (2024) evaluated the noninvasive prenatal genetic “Luna test” using sequence-based copy number analysis of single trophoblasts from maternal blood. Blood samples were collected from 401 pregnant individuals (8-22 weeks), processed to isolate and sequence trophoblast cells. Of the 243 pregnancies undergoing chorionic villus sampling (CVS) or amniocentesis, 160 showed normal results, while 15 had abnormalities, including 14 aneuploidy cases and one Williams syndrome deletion. Placental mosaicism occurred in 7 of 236 Luna test cases and 3 of 188 CVS cases, with no usable trophoblasts recovered in 32 samples. In 158 low-risk pregnancies, 133 were normal, seven showed aneuploidy, and three had likely pathogenic deletions/duplications. Although the sample size was small, the Luna test identified aneuploidies and genetic deletions/duplications, showing consistency with CVS/amniocentesis results, though further data is needed for precise test accuracy (Bellair et al., 2024).
The American College of Obstetrics and Gynecologists (ACOG) and the Society for Maternal-Fetal Medicine (SMFM)
The American College of Obstetrics and Gynecologists and the Society for Maternal-Fetal Medicine offered Recommendations for Clinical Management Guidelines for Obstetricians and Gynecologists on Screening for Aneuploidy.
The following recommendations and conclusions are based on good and consistent scientific evidence (Level A):
- “Prenatal genetic screening (serum screening with or without nuchal translucency [NT] ultrasound or cell-free DNA screening) and diagnostic testing (chorionic villus sampling [CVS] or amniocentesis) options should be discussed and offered to all pregnant [individuals] regardless of maternal age or risk of chromosomal abnormality. After review and discussion, every patient has the right to pursue or decline prenatal genetic screening and diagnostic testing.
- If screening is accepted, patients should have one prenatal screening approach, and should not have multiple screening tests performed simultaneously.
- Cell-free DNA is the most sensitive and specific screening test for the common fetal aneuploidies. Nevertheless, it has the potential for false-positive and false-negative results. Furthermore, cell-free DNA testing is not equivalent to diagnostic testing.
- All patients should be offered a second-trimester ultrasound for fetal structural defects, since these may occur with or without fetal aneuploidy; ideally this is performed between 18 and 22 weeks of gestation (with or without second‐trimester maternal serum alpha‐fetoprotein).
- Patients with a positive screening test result for fetal aneuploidy should undergo genetic counseling and a comprehensive ultrasound evaluation with an opportunity for diagnostic testing to confirm results.
- Patients with a negative screening test result should be made aware that this substantially decreases their risk of the targeted aneuploidy but does not ensure that the fetus is unaffected. The potential for a fetus to be affected by genetic disorders that are not evaluated by the screening or diagnostic test should also be reviewed. Even if patients have a negative screening test result, they may choose diagnostic testing later in pregnancy, particularly if additional findings such as fetal anomalies identified on ultrasound examination become evident.
- Patients whose cell-free DNA screening test results are not reported by the laboratory or are uninterpretable (a no‐call test result) should be informed that test failure is associated with an increased risk of aneuploidy, receive further genetic counseling and be offered comprehensive ultrasound evaluation and diagnostic testing.
- If an enlarged nuchal translucency or an anomaly is identified on ultrasound examination, the patient should be offered genetic counseling and diagnostic testing for genetic conditions as well as a comprehensive ultrasound evaluation including detailed ultrasonography at 18 – 22 weeks of gestation to assess for structural abnormalities.
- [Individuals] with a positive screening test result for fetal aneuploidy should be offered further detailed counseling and testing” (ACOG, 2020).
The following recommendations and conclusions are based on limited or inconsistent scientific evidence (Level B):
- “The use of cell-free DNA screening as follow-up for patients with a screen positive serum analyte screening test result is an option for patients who want to avoid a diagnostic test. However, patients should be informed that this approach may delay definitive diagnosis and will fail to identify some fetuses with chromosomal abnormalities.
- In clinical situations of an isolated soft ultrasonographic marker (such as echogenic cardiac focus, choroid plexus cyst, pyelectasis, short humerus or femur length) where aneuploidy screening has not been performed, the patient should be counseled regarding the risk of aneuploidy associated with the finding and cell-free DNA, quad screen testing, or amniocentesis should be offered. If aneuploidy testing is performed and is low-risk, then no further risk assessment is needed. If more than one marker is identified, then genetic counseling, maternal–fetal medicine consultation, or both are recommended.
- No method of aneuploidy screening that includes a serum sample is as accurate in twin gestations as it is in singleton pregnancies; this information should be incorporated into pretest counseling for patients with multiple gestations.
- Cell-free DNA screening can be performed in twin pregnancies. Overall, performance of screening for trisomy 21 by cell-free DNA in twin pregnancies is encouraging, but the total number of reported affected cases is small. Given the small number of affected cases it is difficult to determine an accurate detection rate for trisomy 18 and 13” (ACOG, 2020).
The following recommendations and conclusions are based primarily on consensus and expert opinion (Level C):
- “The use of multiple serum screening approaches performed independently (eg, a first-trimester screening test followed by a quad screen as an unlinked test) is not recommended because it will result in an unacceptably high positive screening rate and could deliver contradictory risk estimates.
- In multifetal gestations, if a fetal demise, vanishing twin, or anomaly is identified in one fetus, there is a significant risk of an inaccurate test result if serum-based aneuploidy screening or cell-free DNA is used. This information should be reviewed with the patient and diagnostic testing should be offered.
- Patients with unusual or multiple aneuploidies detected by cell-free DNA should be referred for genetic counseling and maternal–fetal medicine consultation” (ACOG, 2020).
The American College of Obstetrics and Gynecologists also comments on the specific types of screening, which include triple, quadruple (quad) and “penta” screens. These screens may be performed in the first trimester (10-14 weeks gestation) or second trimester (15-22 weeks). Triple screens measure serum hCG [human chorionic gonadotropin], AFP [alphafetoprotein], and uE3 [unconjugated estriol], while the quad screen includes DIA [dimeric inhibin A] with the three previously mentioned markers. Some laboratories have been noted to offer a “penta” screen, which includes hyperglycosylated hCG along with the four analytes of the quad screen, but ACOG states that “its performance has not been evaluated rigorously in prospective studies”. ACOG discusses several testing algorithms in the include, which are summarized in the table below:

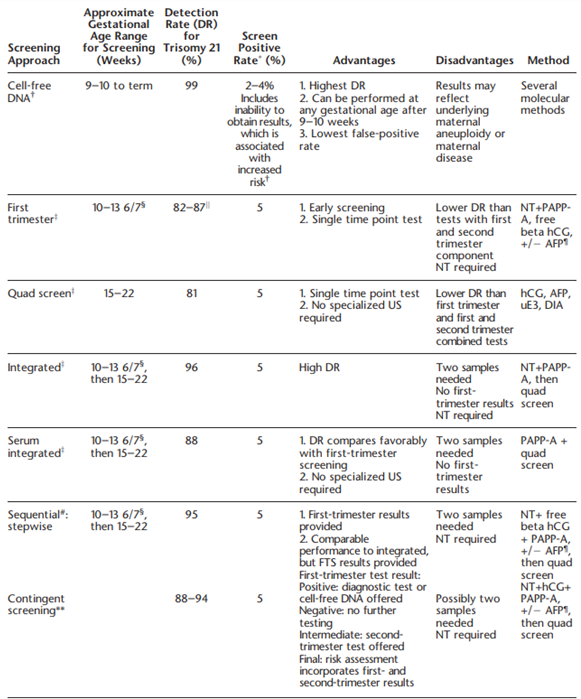
Finally, ACOG notes that other trisomies, such as trisomies 16 or 22, can be tested for. However, ACOG recommends against screening for these two aneuploidies due to lack of validated data (ACOG, 2020).
The National Society of Genetic Counselors (NSGC)
The NSGC issued a position statement that supports noninvasive prenatal screening as an option for pregnancies considered high-risk for trisomy 13, 18 or 21. “The National Society of Genetic Counselors currently supports Noninvasive Prenatal Testing/Noninvasive Prenatal Diagnosis (NIPT/NIPD) as an option for patients whose pregnancies are considered to be at an increased risk for certain chromosome abnormalities. NSGC urges that NIPT/NIPD only be offered in the context of informed consent, education, and counseling by a qualified provider, such as a certified genetic counselor. Patients whose NIPT/NIPD results are abnormal, or who have other factors suggestive of a chromosome abnormality, should receive genetic counseling and be given the option of standard confirmatory diagnostic testing” (Devers et al., 2013).
The NSGC expounded upon their recommendations for prenatal screening and diagnostic testing for chromosomal aneuploidy in a set of practice guidelines. For all patients, it is recommended that “Providers should offer the options of maternal serum screening (MSS) and diagnostic testing for chromosome aneuploidy to every patient”, provided that the providers themselves are made aware of factors that may impact their patients’ options and that the patients are made aware of the costs and benefits of such options. However, “An ultrasound to assess the fetal anatomy is suggested at approximately 18w0d-20w0d gestation for all patients regardless of whether or not they choose to have screening or diagnostic testing” (Wilson et al., 2013).
The NSGC also presented the following recommendations for low-risk patients less than 14 weeks of gestation:
- “For patients who may consider CVS [chorionic villi screening] or amniocentesis, stepwise sequential screening or combined first trimester screening should be considered.”
- “If CVS is not an option, integrated screening may be considered in order to maximize detection rates.”
- “If a patient completes combined first trimester screening, a separate second trimester MSS for chromosome aneuploidy is NOT indicated. Screening for chromosome aneuploidy in the second trimester in patients who present prior to 14 weeks should ONLY be performed as a part of integrated, serum integrated, stepwise sequential, or contingency screening.”
- “Patients who have an increased NT [nuchal translucency] (≥ 95th% or ≥ 3.0 mm) should be offered diagnostic testing by either CVS or amniocentesis. A referral for a fetal echocardiogram should also be considered if the NT ≥ 3.5mm.”
- “Early amniocentesis (prior to 15 weeks of gestation) is not recommended due to the increased risks for pregnancy loss, clubfoot, and fluid leakage. CVS should be offered as the diagnostic testing option for chromosome aneuploidy in the first trimester.”
For low-risk patients after 14 weeks of gestation, they recommend the following:
- “Patients who desire MSS but did not have MSS in the first trimester should be offered a quad or penta screen rather than a triple screen due to the increased detection rates.
- Amniocentesis should be offered as the diagnostic testing option for chromosome aneuploidy for patients after 15 weeks of gestation.”
The NSGC also recommend for those patients at increased risk for chromosome aneuploidy that if the patient presents prior to 14 weeks gestation, “CVS and amniocentesis should both be offered as diagnostic testing options for chromosome aneuploidy”, whereas if the patient presents after 14 weeks gestation, “amniocentesis should be offered as the diagnostic testing option for chromosome aneuploidy.” Lastly, the NSGC reiterated that patients may be offered NIPT (noninvasive prenatal testing) should they desire screening information (Wilson et al., 2013).
The International Society for Prenatal Diagnosis (ISPD)
The ISPD recognizes that massively parallel sequencing for detection of Down syndrome can be helpful for individuals with high-risk pregnancies, when “suitable genetic counseling” is provided (Benn et al., 2013).
The International Society for Prenatal Diagnosis released a statement regarding cell-free DNA screening for Down syndrome in multifetal pregnancies in 2020. The following recommendations were made:
- “The use of first trimester cfDNA screening for the common autosomal trisomies is appropriate for twin pregnancies due to sufficient evidence showing high detection and low false positive rates with high predictive values.
- The finding of an increased risk on a cfDNA screening test in multiple pregnancies should be followed by counseling and an offer of diagnostic testing to confirm results. … CVS and amniocentesis procedures in multiple pregnancies are reliable and safe when performed by a provider experienced in these situations; subsequent diagnostic tests are highly reliable. … Maternal age and nuchal translucency (with or without biochemistry) detects up to 80% of Down syndrome at a 5% false positive rate in twin pregnancies.
- It is preferable for laboratories performing cfDNA testing in multifetal pregnancies to take evidence of zygosity (e.g., chorionicity, sex of the fetuses, embryo transfer history) for the interpretation of both test results and fetal fractions. … Interpretation of the cfDNA test results could differ depending on test methodology, fetal fraction and chorionicity/zygosity. … Fetal fractions are higher in twin pregnancies, but lower for individual fetuses when compared to singletons. Fetal fractions are correlated between dizygous twins, but can still vary two-fold.
- cfDNA based screening for common trisomies in twins provides higher positive predictive values among twin pregnancies compared with traditional serum and nuchal translucency-based screening in twins, but are associated with test failures…When a cfDNA test failure occurs consider ultrasound and diagnostic testing. If there is sufficient time, a second sample draw may also be considered.
- Screening options for triplet pregnancies are lacking and cfDNA may be a potential option. However, diagnostic testing should always be offered and the limitations of screening tests stressed” (Palomaki et al., 2020).
The American College of Medical Genetics and Genomics (ACMG)
The ACMG notes that “Pretest information should be provided … to ensure patients make informed decisions. Aneuploidy screening is not a routine prenatal test; it is acceptable for patients to decline screening.” The ACMG also cautions that “All reports should clearly state that NIPS is a screening test and not diagnostic,” and that results be presented in a clear and easily understandable fashion. ACMG guidelines state that “in pregnancies with multiple gestations and/or donor oocytes, testing laboratories should be contacted regarding the validity of NIPS before it is offered to the patient as a screening option” (Gregg et al., 2016).
The American College of Medical Genetics and Genomics published an evidence-based clinical guideline in 2023 on noninvasive prenatal screening (NIPS) for fetal chromosome abnormalities in a general-risk population. Their recommendations are reported below.
- “Recommendation: ACMG Recommends NIPS Over Traditional Screening Methods for All Pregnant Patients With Singleton Gestation for Fetal Trisomies 21, 18, and 13 (Strong Recommendation Based on High Certainty of Evidence)”
- “Recommendation: ACMG Recommends NIPS Over Traditional Methods for Trisomy Screening in Twin Gestations (Strong Recommendation, Based on High Certainty of Evidence)”
- “Recommendation: ACMG Recommends That NIPS Be Offered to Patients with a singleton gestation to Screen for Fetal SCA [sex chromosome aneuploidies] (Strong Recommendation, Based on High Certainty of Evidence)”
- “Recommendation: ACMG Suggests That NIPS for 22q11.2 Deletion Syndrome Be Offered to All Patients (Conditional Recommendation, Based on Moderate Certainty of the Evidence)”
- “A conditional recommendation should be interpreted as follows: most patients would request this and most clinicians would offer NIPS for this purpose, after a discussion about the benefits and limitations of screening and in the context of shared decision-making.”
- “Recommendation: At This Time, There Is Insufficient Evidence to Recommend Routine Screening for CNVs Other Than 22q11.2 Deletions (No Recommendation, Owing to Lack of Clinically Relevant Evidence and Validation)”
“Recommendation: At This Time, There Is Insufficient Evidence to Recommend or Not Recommend NIPS for the Identification of RATs [rare autosomal trisomies] (No Recommendation, Owing to Lack of Clinically Relevant Evidence)” (Dungan et al., 2023)
Practice Committee and Genetic Counseling Professional Group (GCPG) of the American Society for Reproductive Medicine)
The American Society of Reproductive Medicine notes that screening tests (maternal serum biochemical screening, nuchal translucency and fetal anatomy scan, and noninvasive prenatal screening with cell-free DNA) cannot diagnose chromosomal aneuploidy. “In some cases, ultrasound and biochemical analytes may help identify congenital anomalies that may be associated with an aneuploid pregnancy; however, many aneuploidies (and mosaic aneuploidies in particular) do not result in visible ultrasound anomalies or skewed biochemical analytes and may be easily missed.” Cell-free DNA testing “may test for a select number of full and partial aneuploidies, or all aneuploidies within a specified chromosomal resolution, depending on the specific test used by the laboratory. If the chromosome or chromosomal segment of interest is in fact able to be assessed by the assay used, an aneuploidy may be detected. However, it is important to recognize that NIPT is not designed for the detection of mosaicism and may result in false-negative results. False-positive results may also occur because NIPT analyzes placental (and not fetal) DNA” (ASRM, 2020).
References:
- ACOG. (2015). Committee Opinion No. 640: Cell-Free DNA Screening For Fetal Aneuploidy. Obstet Gynecol, 126(3), e31-37. https://doi.org/10.1097/aog.0000000000001051
- ACOG. (2016). Practice Bulletin No. 163: Screening for Fetal Aneuploidy. Obstet Gynecol, 127(5), e123-137. https://doi.org/10.1097/aog.0000000000001406
- ACOG. (2020). ACOG Practice Bulletin Number 226: Screening for Fetal Chromosomal Abnormalities. American College of Obstetricians and Gynecologists’Committee on Practice Bulletins. https://doi.org/10.1097/AOG.0000000000004084
- AgilentTechnologies. (2022). Clarigo https://www.agilent.com/cs/library/brochures/Clarigo%20patient%20brochure%20v1708-EN.pdf
- Al Toukhi, S., Chitayat, D., Keunen, J., Roifman, M., Seaward, G., Windrim, R., Ryan, G., & Van Mieghem, T. (2019). Impact of introduction of noninvasive prenatal testing on uptake of genetic testing in fetuses with central nervous system anomalies. Prenatal Diagnosis, 39, 544-548. https://doi.org/10.1002/pd.5466
- Aradhya, S., Manning, M. A., Splendore, A., & Cherry, A. M. (2007). Whole-genome array-CGH identifies novel contiguous gene deletions and duplications associated with developmental delay, mental retardation, and dysmorphic features. American Journal of Medical Genetics Part A, 143A(13), 1431-1441. https://doi.org/10.1002/ajmg.a.31773
- ASRM. (2020). Clinical management of mosaic results from preimplantation genetic testing for aneuploidy (PGT-A) of blastocysts: a committee opinion. Fertility and Sterility, 114(2), 246-254. https://doi.org/10.1016/j.fertnstert.2020.05.014
- Baer, R. J., Flessel, M. C., Jelliffe-Pawlowski, L. L., Goldman, S., Hudgins, L., Hull, A. D., Norton, M. E., & Currier, R. J. (2015). Detection Rates for Aneuploidy by First-Trimester and Sequential Screening. 126(4), 753-759. https://doi.org/10.1097/aog.0000000000001040
- Bellair, M., Amaral, E., Ouren, M., Roark, C., Kim, J., O'Connor, A., Soriano, A., Schindler, M. L., Wapner, R. J., Stone, J. L., Tavella, N., Merriam, A., Perley, L., Breman, A. M., & Beaudet, A. L. (2024). Noninvasive single-cell-based prenatal genetic testing: A proof of concept clinical study. Prenatal Diagnosis, 44(3), 304-316. https://doi.org/10.1002/pd.6529
- Benn, P., Borell, A., Chiu, R., Cuckle, H., Dugoff, L., Faas, B., Gross, S., Johnson, J., Maymon, R., Norton, M., Odibo, A., Schielen, P., Spencer, K., Huang, T., Wright, D., & Yaron, Y. (2013). Position statement from the Aneuploidy Screening Committee on behalf of the Board of the International Society for Prenatal Diagnosis. Prenat Diagn, 33(7), 622-629. https://doi.org/10.1002/pd.4139
- BGI. (2023). The NIFTY® Test. https://www.bgi.com/global/molecular-genetics/nifty-non-invasive-prenatal-testing/the-nifty-test-non-invasive-prenatal-testing/
- Bianchi, D., Chudova, D., Sehnert, A., Bhatt, S., Murray, K., Prosen, T., Garber, J., Wilkins-Haug, L., Vora, N., & Warsof, S. (2015). Noninvasive prenatal testing and incidental detection of occult maternal malignancies. JAMA, 314, 162-169. https://doi.org/10.1001/jama.2015.7120
- Bianchi, D. W., Platt, L. D., Goldberg, J. D., Abuhamad, A. Z., Sehnert, A. J., & Rava, R. P. (2012). Genome-wide fetal aneuploidy detection by maternal plasma DNA sequencing. Obstet Gynecol, 119(5), 890-901. https://doi.org/10.1097/AOG.0b013e31824fb482
- Bornstein, E., Gulersen, M., Krantz, D., Cheung, S. W., Maliszewski, K., & Divon, M. Y. (2018). Microarray analysis: First-trimester maternal serum free beta-hCG and the risk of significant copy number variants. Prenat Diagn, 38(12), 971-978. https://doi.org/10.1002/pd.5350
- Centogene. (2023). CentoNIPT®. https://www.centogene.com/diagnostics/our-tests/non-invasive-prenatal-testing/
- Dai, P., Yang, Y., Zhao, G., Gu, Z., Ren, H., Hu, S., Liu, N., Jiao, W., Li, J., & Kong, X. (2022). A dPCR-NIPT assay for detections of trisomies 21, 18 and 13 in a single-tube reaction-could it replace serum biochemical tests as a primary maternal plasma screening tool? J Transl Med, 20(1), 269. https://doi.org/10.1186/s12967-022-03455-y
- Dar, P., Jacobsson, B., MacPherson, C., Egbert, M., Malone, F., Wapner, R. J., Roman, A. S., Khalil, A., Faro, R., Madankumar, R., Edwards, L., Haeri, S., Silver, R., Vohra, N., Hyett, J., Clunie, G., Demko, Z., Martin, K., Rabinowitz, M., . . . Norton, M. E. (2022). Cell-free DNA screening for trisomies 21, 18, and 13 in pregnancies at low and high risk for aneuploidy with genetic confirmation. Am J Obstet Gynecol, 227(2), 259.e251-259.e214. https://doi.org/10.1016/j.ajog.2022.01.019
- Devers, P. L., Cronister, A., Ormond, K. E., Facio, F., Brasington, C. K., & Flodman, P. (2013). Noninvasive prenatal testing/noninvasive prenatal diagnosis: the position of the National Society of Genetic Counselors. J Genet Couns, 22(3), 291-295. https://doi.org/10.1007/s10897-012-9564-0
- Dey, M., Sharma, S., & Aggarwal, S. (2013). Prenatal screening methods for aneuploidies. N Am J Med Sci, 5(3), 182-190. https://pubmed.ncbi.nlm.nih.gov/23626953/
- Dungan, J. S., Klugman, S., Darilek, S., Malinowski, J., Akkari, Y. M. N., Monaghan, K. G., Erwin, A., & Best, R. G. (2023). Noninvasive prenatal screening (NIPS) for fetal chromosome abnormalities in a general-risk population: An evidence-based clinical guideline of the American College of Medical Genetics and Genomics (ACMG). Genet Med, 25(2), 100336. https://doi.org/10.1016/j.gim.2022.11.004
- Eiben, B., Krapp, M., Borth, H., Kutur, N., Kreiselmaier, P., Glaubitz, R., Deutinger, J., & Merz, E. (2015). Single Nucleotide Polymorphism-Based Analysis of Cell-Free Fetal DNA in 3000 Cases from Germany and Austria. Ultrasound Int Open, 1(1), E8-e11. https://doi.org/10.1055/s-0035-1555765
- FDA. (2022). Genetic Non-Invasive Prenatal Screening Tests May Have False Results: FDA Safety Communication. https://www.fda.gov/medical-devices/safety-communications/genetic-non-invasive-prenatal-screening-tests-may-have-false-results-fda-safety-communication?
- Futch, T., Spinosa, J., Bhatt, S., de Feo, E., Rava, R. P., & Sehnert, A. J. (2013). Initial clinical laboratory experience in noninvasive prenatal testing for fetal aneuploidy from maternal plasma DNA samples. Prenat Diagn, 33(6), 569-574. https://doi.org/10.1002/pd.4123
- Garshasbi, M., Wang, Y., Hantoosh Zadeh, S., Giti, S., Piri, S., & Reza Hekmat, M. (2019). Clinical Application of Cell-Free DNA Sequencing-Based Noninvasive Prenatal Testing for Trisomies 21, 18, 13 and Sex Chromosome Aneuploidy in a Mixed-Risk Population in Iran. Fetal Diagn Ther, 1-8. https://doi.org/10.1159/000501014
- Genetics, I. (2016). informaSeq® noninvasive prenatal test. Illumina, Inc. https://www.labcorp.com/tests/related-documents/L13272
- GenPath. (2023). CLARITESTTM CORE NON-INVASIVE PRENATAL SCREENING. https://www.genpathdiagnostics.com/hcp/womens-health/prenatal-screening/claritest/
- Glenn E Palomaki, P. M. M., PhDJacquelyn V Halliday, MS. (2024, June 19, 2024). Prenatal screening for common aneuploidies using cell-free DNA. https://www.uptodate.com/contents/prenatal-screening-for-common-aneuploidies-using-cell-free-dna?
- Gregg, A. R., Skotko, B. G., Benkendorf, J. L., Monaghan, K. G., Bajaj, K., Best, R. G., Klugman, S., & Watson, M. S. (2016). Noninvasive prenatal screening for fetal aneuploidy, 2016 update: a position statement of the American College of Medical Genetics and Genomics. Genet Med, 18(10), 1056-1065. https://doi.org/10.1038/gim.2016.97
- Heesterbeek, C. J., Aukema, S. M., Galjaard, R.-J. H., Boon, E. M. J., Srebniak, M. I., Bouman, K., Faas, B. H. W., Govaerts, L. C. P., Hoffer, M. J. V., den Hollander, N. S., Lichtenbelt, K. D., van Maarle, M. C., van Prooyen Schuurman, L., van Rij, M. C., Schuring-Blom, G. H., Stevens, S. J. C., Tan-Sindhunata, G., Zamani Esteki, M., de Die-Smulders, C. E. M., . . . Macville, M. V. E. (2022). Noninvasive Prenatal Test Results Indicative of Maternal Malignancies: A Nationwide Genetic and Clinical Follow-Up Study. Journal of Clinical Oncology, 40(22), 2426-2435. https://doi.org/10.1200/JCO.21.02260
- Illumina. (2023). Verifi Prenatal Test Services. https://www.illumina.com/clinical/illumina_clinical_laboratory/verifi-prenatal-tests.html
- Invitae. (2023). Invitae Non-invasive Prenatal Screening. https://www.invitae.com/en/noninvasive-prenatal-screening/
- ISPD. (2018). Joint Position Statement from the International Society for Prenatal Diagnosis (ISPD), the Society for Maternal Fetal Medicine (SMFM), and the Perinatal Quality Foundation (PQF) on the use of genome-wide sequencing for fetal diagnosis. Prenat Diagn, 38(1), 6-9. https://doi.org/10.1002/pd.5195
- LabCorp. (2023a). Early risk assessment of Down syndrome and other conditions. Retrieved 10/10/2022 from https://www.integratedgenetics.com/patients/pregnancy/maternit21plus
- LabCorp. (2023b). First and second trimester serum screening. Laboratory Corporation of America Holdings. Retrieved 11/11 from https://www.integratedgenetics.com/patients/pregnancy/serum-screening
- Lee, Chen, L. C., Cheong, M. L., Chou, C. Y., & Tsai, M. S. (2013). First trimester combined test for Down syndrome screening in unselected pregnancies - a report of a 13-year experience. Taiwan J Obstet Gynecol, 52(4), 523-526. https://doi.org/10.1016/j.tjog.2013.10.012
- Lee, Kim, H., Park, J., Yun, T., Park, D. Y., Kim, M., & Ryu, H. M. (2019). Clinical Validation of Non-Invasive Prenatal Testing for Fetal Common Aneuploidies in 1,055 Korean Pregnant Women: a Single Center Experience. J Korean Med Sci, 34(24), e172. https://doi.org/10.3346/jkms.2019.34.e172
- Liang, D., Cram, D. S., Tan, H., Linpeng, S., Liu, Y., Sun, H., Zhang, Y., Tian, F., Zhu, H., Xu, M., Wang, H., Yu, F., & Wu, L. (2019). Clinical utility of noninvasive prenatal screening for expanded chromosome disease syndromes. Genetics in Medicine, 21(9), 1998-2006. https://doi.org/10.1038/s41436-019-0467-4
- Lo, Y. M., Corbetta, N., Chamberlain, P. F., Rai, V., Sargent, I. L., Redman, C. W., & Wainscoat, J. S. (1997). Presence of fetal DNA in maternal plasma and serum. Lancet, 350(9076), 485-487. https://doi.org/10.1016/s0140-6736(97)02174-0
- Luna. (2022a). Luna Prenatal Test. Luna Genetics. https://cdn.prod.website-files.com/60adc548ba98e3382beed23d/62a91215f4f2bf0f6bbe361f_QSE8.FORM.004%20Luna%20Prenatal%20Test%20Information%20Sheet%20v2.0.pdf
- Luna. (2022b). Luna Prenatal Test White Paper. https://assets.website-files.com/60adc548ba98e3382beed23d/62a915e6d02dc869709d0bea_QSE8.FORM.005%20Luna%20Prenatal%20Test%20White%20Paper%20v2.0.pdf
- Luo, Y., Hu, H., Zhang, R., Ma, Y., Pan, Y., Long, Y., Hu, B., Yao, H., & Liang, Z. (2021). An assessment of the analytical performance of non-invasive prenatal testing (NIPT) in detecting sex chromosome aneuploidies: 34,717-patient sample in a single prenatal diagnosis Centre in China. J Gene Med, 23(9), e3362. https://doi.org/10.1002/jgm.3362
- Malone, F. D., Canick, J. A., Ball, R. H., Nyberg, D. A., Comstock, C. H., Bukowski, R., Berkowitz, R. L., Gross, S. J., Dugoff, L., Craigo, S. D., Timor-Tritsch, I. E., Carr, S. R., Wolfe, H. M., Dukes, K., Bianchi, D. W., Rudnicka, A. R., Hackshaw, A. K., Lambert-Messerlian, G., Wald, N. J., & D'Alton, M. E. (2005). First-trimester or second-trimester screening, or both, for Down's syndrome. N Engl J Med, 353(19), 2001-2011. https://doi.org/10.1056/NEJMoa043693
- McKanna, T., Ryan, A., Krinshpun, S., Kareht, S., Marchand, K., Grabarits, C., Ali, M., McElheny, A., Gardiner, K., LeChien, K., Hsu, M., Saltzman, D., Stosic, M., Martin, K., & Benn, P. (2018). Fetal fraction-based risk algorithm for non-invasive prenatal testing: screening for trisomy 13, 18, and triploidy in women with low cell-free fetal DNA. Ultrasound Obstet Gynecol. https://doi.org/10.1002/uog.19176
- Muzzey, D., Goldberg, J. D., & Haverty, C. (2020). Noninvasive prenatal screening for patients with high body mass index: Evaluating the impact of a customized whole genome sequencing workflow on sensitivity and residual risk. Prenat Diagn, 40(3), 333-341. https://doi.org/10.1002/pd.5603
- Myriad. (2023). MYRIAD PREQUELTM PRENATAL SCREEN. https://myriadwomenshealth.com/patient-prequel/
- Natera. (2023). Panorama Test. https://www.natera.com/panorama-test
- Neufeld-Kaiser, W. A., Cheng, E. Y., & Liu, Y. J. (2015). Positive predictive value of non-invasive prenatal screening for fetal chromosome disorders using cell-free DNA in maternal serum: independent clinical experience of a tertiary referral center. BMC Med, 13, 129. https://doi.org/10.1186/s12916-015-0374-8
- NIPD. (2023, 2021). NIPD Genetics VERACITY new generation NIPT. NIPD Genetics. https://nipd.com/products/prenatal/veracity-patients/
- Norton, M. E., Jacobsson, B., Swamy, G. K., Laurent, L. C., Ranzini, A. C., Brar, H., Tomlinson, M. W., Pereira, L., Spitz, J. L., Hollemon, D., Cuckle, H., Musci, T. J., & Wapner, R. J. (2015). Cell-free DNA analysis for noninvasive examination of trisomy. N Engl J Med, 372(17), 1589-1597. https://doi.org/10.1056/NEJMoa1407349
- Palomaki, G. E., Chiu, R. W. K., Pertile, M. D., Sistermans, E. A., Yaron, Y., Vermeesch, J. R., Vora, N. L., Best, R. G., & Wilkins-Haug, L. (2020). International Society for Prenatal Diagnosis (ISPD) Position Statement: Cell free (cf)DNA screening for Down syndrome in multiple pregnancies. Prenatal Diagnosis, Accepted for publication(n/a). https://doi.org/10.1002/pd.5832
- Palomaki, G. E., Deciu, C., Kloza, E. M., Lambert-Messerlian, G. M., Haddow, J. E., Neveux, L. M., Ehrich, M., van den Boom, D., Bombard, A. T., Grody, W. W., Nelson, S. F., & Canick, J. A. (2012). DNA sequencing of maternal plasma reliably identifies trisomy 18 and trisomy 13 as well as Down syndrome: an international collaborative study. Genet Med, 14(3), 296-305. https://doi.org/10.1038/gim.2011.73
- Palomaki, G. E., Kloza, E. M., Lambert-Messerlian, G. M., Haddow, J. E., Neveux, L. M., Ehrich, M., van den Boom, D., Bombard, A. T., Deciu, C., Grody, W. W., Nelson, S. F., & Canick, J. A. (2011). DNA sequencing of maternal plasma to detect Down syndrome: an international clinical validation study. Genet Med, 13(11), 913-920. https://doi.org/10.1097/GIM.0b013e3182368a0e
- Palomaki, G. E., Messerlian, G. M., & Halliday, J. V. (2024, Jun 19, 2024). Prenatal screening for common aneuploidies using cell-free DNA. https://www.uptodate.com/contents/prenatal-screening-for-common-aneuploidies-using-cell-free-dna
- Palomaki, G. E., Neveux, L. M., Knight, G. J., Haddow, J. E., & Pandian, R. (2004). Maternal Serum Invasive Trophoblast Antigen (Hyperglycosylated hCG) as a Screening Marker for Down Syndrome during the Second Trimester. Clinical Chemistry, 50(10), 1804-1808. https://doi.org/10.1373/clinchem.2004.038059
- Park, S. Y., Jang, I. A., Lee, M. A., Kim, Y. J., Chun, S. H., & Park, M. H. (2016). Screening for chromosomal abnormalities using combined test in the first trimester of pregnancy. Obstet Gynecol Sci, 59(5), 357-366. https://doi.org/10.5468/ogs.2016.59.5.357
- PerkinElmer. (2023). VANADIS® NIPT SYSTEM A Whole New Way to NIPT. https://rh.perkinelmer.com/products/vanadis-nipt-system/
- QuestDiagnostics. (2022). Prenatal Screening and Diagnosis of Neural Tube Defects, Down Syndrome, Trisomy 18, and Trisomy 13. https://testdirectory.questdiagnostics.com/test/test-guides/TS_Quad_Screen/quad-screen
- QuestDiagnostics. (2023). QNatal® Advanced. http://education.questdiagnostics.com/faq/FAQ167
- Reddy, U. M., Page, G. P., Saade, G. R., Silver, R. M., Thorsten, V. R., Parker, C. B., Pinar, H., Willinger, M., Stoll, B. J., Heim-Hall, J., Varner, M. W., Goldenberg, R. L., Bukowski, R., Wapner, R. J., Drews-Botsch, C. D., O'Brien, B. M., Dudley, D. J., & Levy, B. (2012). Karyotype versus Microarray Testing for Genetic Abnormalities after Stillbirth. New England Journal of Medicine, 367(23), 2185-2193. https://doi.org/10.1056/NEJMoa1201569
- Robson, S. C., Chitty, L. S., Morris, S., Verhoef, T., Ambler, G., Wellesley, D. G., Graham, R., Leader, C., Fisher, J., & Crolla, J. A. (2017). Efficacy and Mechanism Evaluation. In Evaluation of Array Comparative genomic Hybridisation in prenatal diagnosis of fetal anomalies: a multicentre cohort study with cost analysis and assessment of patient, health professional and commissioner preferences for array comparative genomic hybridisation. NIHR Journals Library. Copyright © Queen’s Printer and Controller of HMSO 2017. . https://doi.org/10.3310/eme04010
- Roche. (2023). Harmony Prenatal Test. Retrieved 10/10/2022 from https://harmonytest.roche.com/global/en/home.html
- Shaw, J., Scotchman, E., Chandler, N., & Chitty, L. S. (2020). PREIMPLANTATION GENETIC TESTING: Non-invasive prenatal testing for aneuploidy, copy-number variants and single-gene disorders. Reproduction, 160(5), A1-A11. https://doi.org/10.1530/rep-19-0591
- Shiefa, S., Amargandhi, M., Bhupendra, J., Moulali, S., & Kristine, T. (2013). First Trimester Maternal Serum Screening Using Biochemical Markers PAPP-A and Free β-hCG for Down Syndrome, Patau Syndrome and Edward Syndrome. Indian J Clin Biochem, 28(1), 3-12. https://doi.org/10.1007/s12291-012-0269-9
- Strom, C. M., Anderson, B., Tsao, D., Zhang, K., Liu, Y., Livingston, K., Elzinga, C., Evans, M., Nguyen, Q., Wolfson, D., Rowland, C., Kolacki, P., Maxwell, M., Wang, J. C., Rabin, D., Catanese, J., Owen, R., Braastad, C., & Sun, W. (2017). Improving the Positive Predictive Value of Non-Invasive Prenatal Screening (NIPS). PLoS One, 12(3), e0167130. https://doi.org/10.1371/journal.pone.0167130
- Taneja, P. A., Snyder, H. L., de Feo, E., Kruglyak, K. M., Halks-Miller, M., Curnow, K. J., & Bhatt, S. (2016). Noninvasive prenatal testing in the general obstetric population: clinical performance and counseling considerations in over 85 000 cases. Prenat Diagn, 36(3), 237-243. https://doi.org/10.1002/pd.4766
- van der Meij, K. R. M., Sistermans, E. A., Macville, M. V. E., Stevens, S. J. C., Bax, C. J., Bekker, M. N., Bilardo, C. M., Boon, E. M. J., Boter, M., Diderich, K. E. M., de Die-Smulders, C. E. M., Duin, L. K., Faas, B. H. W., Feenstra, I., Haak, M. C., Hoffer, M. J. V., den Hollander, N. S., Hollink, I. H. I. M., Jehee, F. S., . . . Weiss, M. M. (2019). TRIDENT-2: National Implementation of Genome-wide Non-invasive Prenatal Testing as a First-Tier Screening Test in the Netherlands. The American Journal of Human Genetics, 105(6), 1091-1101. https://doi.org/10.1016/j.ajhg.2019.10.005
- Wald, N. J., Huttly, W. J., Murphy, K. W., Ali, K., Bestwick, J. P., & Rodeck, C. H. (2009). Antenatal screening for Down's syndrome using the Integrated test at two London hospitals. J Med Screen, 16(1), 7-10. https://doi.org/10.1258/jms.2009.008094
- Wald, N. J., Rodeck, C., Hackshaw, A. K., Walters, J., Chitty, L., & Mackinson, A. M. (2003). First and second trimester antenatal screening for Down's syndrome: the results of the Serum, Urine and Ultrasound Screening Study (SURUSS). J Med Screen, 10(2), 56-104. https://doi.org/10.1258/096914103321824133
- Westerfield, L., Darilek, S., & van den Veyver, I. B. (2014). Counseling Challenges with Variants of Uncertain Significance and Incidental Findings in Prenatal Genetic Screening and Diagnosis. J Clin Med, 3(3), 1018-1032. https://doi.org/10.3390/jcm3031018
- Wilson, K. L., Czerwinski, J. L., Hoskovec, J. M., Noblin, S. J., Sullivan, C. M., Harbison, A., Campion, M. W., Devary, K., Devers, P., & Singletary, C. N. (2013). NSGC practice guideline: prenatal screening and diagnostic testing options for chromosome aneuploidy. J Genet Couns, 22(1), 4-15. https://doi.org/10.1007/s10897-012-9545-3
- Witters, G., Van Robays, J., Willekes, C., Coumans, A., Peeters, H., Gyselaers, W., & Fryns, J. P. (2011). Trisomy 13, 18, 21, Triploidy and Turner syndrome: the 5T's. Look at the hands. Facts Views Vis Obgyn, 3(1), 15-21. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3991414/
- Wu, X., Li, Y., Xie, X., Su, L., Cai, M., Lin, N., Du, S., Xu, L., & Huang, H. (2020). Clinical Review of Noninvasive Prenatal Testing: Experience from 551 Pregnancies with Noninvasive Prenatal Testing–Positive Results in a Tertiary Referral Center. The Journal of Molecular Diagnostics. https://doi.org/10.1016/j.jmoldx.2020.09.008
- YourgeneHealth. (2023). Non-Invasive Prenatal Testing (NIPT). https://www.yourgene-health.com/yourgeneproducts/nipt
Coding Section
| Codes | Number | Description |
| CPT | 81420 | Fetal chromosomal aneuploidy (e.g., trisomy 21, monosomy X) genomic sequence analysis panel, circulating cell-free fetal DNA in maternal blood, must include analysis of chromosomes 13, 18, and 21 |
| 81422 | Fetal chromosomal microdeletion(s) genomic sequence analysis (e.g., DiGeorge syndrome, Cri-du-chat syndrome), circulating cell-free fetal DNA in maternal blood | |
| 81479 | Unlisted molecular pathology procedure | |
| 81507 | Fetal aneuploidy (trisomy 21, 18, and 13) DNA sequence analysis of selected regions using maternal plasma, algorithm reported as a risk score for each trisomy | |
| 81508 | Fetal congenital abnormalities, biochemical assays of two proteins (PAPP-A, hCG), utilizing maternal serum, algorithm reported as risk score | |
| 81509 | Fetal congenital abnormalities, biochemical assays of three proteins (PAPP-A, hCG, DIA), utilizing maternal serum, algorithm reported as risk score | |
| 81510 | Fetal congenital abnormalities, biochemical assays of three analytes (AFP, uE3, hCG), utilizing maternal serum, algorithm reported as risk score | |
| 81511 | Fetal congenital abnormatlities, biochemical assays of four analytes (AFP, uE3, hCG [any form], DIA), utilizing maternal serum, algorithm reported as a risk score (may include additional results from previous biochemical testing) | |
| 81512 | Fetal congenital abnormatlities, biochemical assays of four analytes (AFP, uE3, hCG (any form), DIA), utilizing maternal serum, algorithm reported as a risk score (may include additional results from previous biochemical testing) | |
| 81599 | Unlisted multianalyte assay with algorithmic analysis | |
| 82105 | Alpha-fetoprotein; serum | |
| 82106 | Alpha-fetoprotein; amniotic fluid | |
| 82677 | Estriol | |
| 84163 | Pregnancy-associated plasma protein (PAPP-A) | |
| 84702 | Gonadotropin, chorionic, quantitative | |
| 84703 | qualitative | |
| 84704 | free beta chain | |
| 86336 | Inhibin A | |
| 88235 | Tissue culture for non-neoplastic disorders; amniotic fluid or chorionic villus cells | |
| 88267 | Chromosome analysis; amniotic fluid or chorionic villus, count 15 cells, 1 karyotype, with banding |
|
| 88269 | Chromosome analysis; in Situ for amniotic fluid cells, count cells from 6-12 colonies, 1 karyotype, with banding |
|
| 88271 | Molecular genetics; DNA probe, each (e.g., FISH) |
|
| 88280 | Chromosome analysis; additional karyotypes, each study | |
| 88285 | Chromosome analysis; additional cells counted, each study | |
| 0327U | Fetal aneuploidy (trisomy 13, 18 and 21), DNA sequence analysis of selected regions using maternal plasma. Algorithm reported as a risk score for each trisomy. Includes sex reporting, if performed | |
| 0341U | Fetal Aneuploidy DNA sequencing comparative analysis, Fetal DNA from products of conception, reported as normal (euploidy), monosomy, trisomy, or partial deletion/duplication, mosaicism, and segmental aneuploid |
|
| HCPCS | ||
| ICD-10-CM (effective 10/01/12) | O09.00 – O82 |
Supervision of high-risk pregnancy |
| O88.011 – O88.83 | Obstetric embolism | |
| O91.011 – O91.219 | Infections of breast associated with pregnancy, the puerperium and lactation | |
| O92.011 – O92.13 | Other disorders of breast and disorders of lactation associated with pregnancy and the puerperium | |
| O94 – O9A.53 | Other obstetric conditions, not elsewhere classified | |
| Z33.1 – Z33.3 | Pregnant state | |
| Z3A.00 – Z3A.49 | Weeks of Gestation | |
| Z34.00 – Z34.93 | Encounter for supervision of normal pregnancy | |
| ICD-10-PCS (effective 10/01/15) | ICD-10-PCS codes are only for use on inpatient services. There is no specific ICD-10-PCS code for this testing. |
Procedure and diagnosis codes on Medical Policy documents are included only as a general reference tool for each policy. They may not be all-inclusive.
This medical policy was developed through consideration of peer-reviewed medical literature generally recognized by the relevant medical community, U.S. FDA approval status, nationally accepted standards of medical practice and accepted standards of medical practice in this community, Blue Cross Blue Shield Association technology assessment program (TEC) and other nonaffiliated technology evaluation centers, reference to federal regulations, other plan medical policies, and accredited national guidelines.
"Current Procedural Terminology © American Medical Association. All Rights Reserved"
History From 2024 Forward